Improving CLIP-seq data analysis by incorporating transcript information
- PMID: 33334306
- PMCID: PMC7745353
- DOI: 10.1186/s12864-020-07297-0
Improving CLIP-seq data analysis by incorporating transcript information
Abstract
Background: Current peak callers for identifying RNA-binding protein (RBP) binding sites from CLIP-seq data take into account genomic read profiles, but they ignore the underlying transcript information, that is information regarding splicing events. So far, there are no studies available that closer observe this issue.
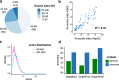
Results: Here we show that current peak callers are susceptible to false peak calling near exon borders. We quantify its extent in publicly available datasets, which turns out to be substantial. By providing a tool called CLIPcontext for automatic transcript and genomic context sequence extraction, we further demonstrate that context choice affects the performances of RBP binding site prediction tools. Moreover, we show that known motifs of exon-binding RBPs are often enriched in transcript context sites, which should enable the recovery of more authentic binding sites. Finally, we discuss possible strategies on how to integrate transcript information into future workflows.
Conclusions: Our results demonstrate the importance of incorporating transcript information in CLIP-seq data analysis. Taking advantage of the underlying transcript information should therefore become an integral part of future peak calling and downstream analysis tools.
Keywords: CLIP-seq; Peak calling; RBP binding site prediction; eCLIP.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

