Attenuation of SRC Kinase Activity Augments PARP Inhibitor-mediated Synthetic Lethality in BRCA2-altered Prostate Tumors
- PMID: 33334906
- PMCID: PMC7956224
- DOI: 10.1158/1078-0432.CCR-20-2483
Attenuation of SRC Kinase Activity Augments PARP Inhibitor-mediated Synthetic Lethality in BRCA2-altered Prostate Tumors
Abstract
Purpose: Alterations in DNA damage repair (DDR) pathway genes occur in 20%-25% of men with metastatic castration-resistant prostate cancer (mCRPC). Although PARP inhibitors (PARPis) have been shown to benefit men with mCRPC harboring DDR defects due to mutations in BRCA1/2 and ATM, additional treatments are necessary because the effects are not durable.
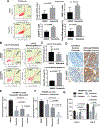
Experimental design: We performed transcriptomic analysis of publicly available mCRPC cases, comparing BRCA2 null with BRCA2 wild-type. We generated BRCA2-null prostate cancer cells using CRISPR/Cas9 and treated these cells with PARPis and SRC inhibitors. We also assessed the antiproliferative effects of combination treatment in 3D prostate cancer organoids.
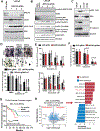
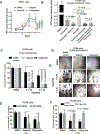
Results: We observed significant enrichment of the SRC signaling pathway in BRCA2-altered mCRPC. BRCA2-null prostate cancer cell lines had increased SRC phosphorylation and higher sensitivity to SRC inhibitors (e.g., dasatinib, bosutinib, and saracatinib) relative to wild-type cells. Combination treatment with PARPis and SRC inhibitors was antiproliferative and had a synergistic effect in BRCA2-null prostate cancer cells, mCRPC organoids, and Trp53/Rb1-null prostate cancer cells. Inhibition of SRC signaling by dasatinib augmented DNA damage in BRCA2-null prostate cancer cells. Moreover, SRC knockdown increased PARPi sensitivity in BRCA2-null prostate cancer cells.
Conclusions: This work suggests that SRC activation may be a potential mechanism of PARPi resistance and that treatment with SRC inhibitors may overcome this resistance. Our preclinical study demonstrates that combining PARPis and SRC inhibitors may be a promising therapeutic strategy for patients with BRCA2-null mCRPC.
©2020 American Association for Cancer Research.
Conflict of interest statement
The other authors have no potential conflicts of interest to disclose.
Figures



Similar articles
-
Differential Activity of PARP Inhibitors in BRCA1- Versus BRCA2-Altered Metastatic Castration-Resistant Prostate Cancer.JCO Precis Oncol. 2021 Jul 22;5:PO.21.00070. doi: 10.1200/PO.21.00070. eCollection 2021. JCO Precis Oncol. 2021. PMID: 34778690 Free PMC article.
-
Androgen receptor inhibitor-induced "BRCAness" and PARP inhibition are synthetically lethal for castration-resistant prostate cancer.Sci Signal. 2017 May 23;10(480):eaam7479. doi: 10.1126/scisignal.aam7479. Sci Signal. 2017. PMID: 28536297 Free PMC article.
-
Differential responses to taxanes and PARP inhibitors in ATM- versus BRCA2-mutated metastatic castrate-resistant prostate cancer.Prostate. 2023 Feb;83(3):227-236. doi: 10.1002/pros.24454. Epub 2022 Nov 16. Prostate. 2023. PMID: 36382533 Free PMC article.
-
DNA-Damage-Repair Gene Alterations in Genitourinary Malignancies.Eur Surg Res. 2022;63(4):155-164. doi: 10.1159/000526415. Epub 2022 Aug 9. Eur Surg Res. 2022. PMID: 35944490 Review.
-
Is It Time to Anticipate the Use of PARP Inhibition in Prostate Cancer Patients?Curr Oncol. 2023 Aug 30;30(9):8054-8067. doi: 10.3390/curroncol30090584. Curr Oncol. 2023. PMID: 37754499 Free PMC article. Review.
Cited by
-
Establishment of a novel prognostic prediction model through bioinformatics analysis for prostate cancer based on ferroptosis-related genes and its application in immune cell infiltration.Transl Androl Urol. 2022 Aug;11(8):1130-1147. doi: 10.21037/tau-22-454. Transl Androl Urol. 2022. PMID: 36092848 Free PMC article.
-
Targeting mRNA-coding genes in prostate cancer using CRISPR/Cas9 technology with a special focus on androgen receptor signaling.Cell Commun Signal. 2024 Oct 17;22(1):504. doi: 10.1186/s12964-024-01833-1. Cell Commun Signal. 2024. PMID: 39420406 Free PMC article. Review.
-
pH-Responsive Hyaluronic Acid Nanoparticles for Enhanced Triple Negative Breast Cancer Therapy.Int J Nanomedicine. 2022 Mar 25;17:1437-1457. doi: 10.2147/IJN.S360500. eCollection 2022. Int J Nanomedicine. 2022. PMID: 35369031 Free PMC article.
-
BCL2 drives castration resistance in castration-sensitive prostate cancer by orchestrating reciprocal crosstalk between oncogenic pathways.Cell Rep. 2025 Jun 24;44(6):115779. doi: 10.1016/j.celrep.2025.115779. Epub 2025 May 30. Cell Rep. 2025. PMID: 40448998 Free PMC article.
-
A Conformation Selective Mode of Inhibiting SRC Improves Drug Efficacy and Tolerability.Cancer Res. 2021 Nov 1;81(21):5438-5450. doi: 10.1158/0008-5472.CAN-21-0613. Epub 2021 Aug 20. Cancer Res. 2021. PMID: 34417202 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

