Phylogenetic comparisons reveal mosaic histories of larval and adult shell matrix protein deployment in pteriomorph bivalves
- PMID: 33335265
- PMCID: PMC7747718
- DOI: 10.1038/s41598-020-79330-x
Phylogenetic comparisons reveal mosaic histories of larval and adult shell matrix protein deployment in pteriomorph bivalves
Abstract
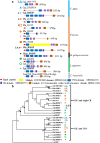
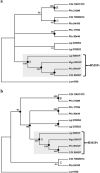
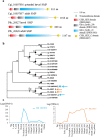
Molluscan shells are organo-mineral composites, in which the dominant calcium carbonate is intimately associated with an organic matrix comprised mainly of proteins and polysaccharides. However, whether the various shell matrix proteins (SMPs) date to the origin of hard skeletons in the Cambrian, or whether they represent later deployment through adaptive evolution, is still debated. In order to address this issue and to better understand the origins and evolution of biomineralization, phylogenetic analyses have been performed on the three SMP families, Von Willebrand factor type A (VWA) and chitin-binding domain-containing protein (VWA-CB dcp), chitobiase, and carbonic anhydrase (CA), which exist in both larval and adult shell proteomes in the bivalves, Crassostrea gigas and Pinctada fucata. In VWA-CB dcp and chitobiase, paralogs for larval and adult SMPs evolved before the divergence of these species. CA-SMPs have been taken as evidence for ancient origins of SMPs by their presumed indispensable function in biomineralization and ubiquitous distribution in molluscs. However, our results indicate gene duplications that gave rise to separate deployments as larval and adult CA-SMPs occurred independently in each lineage after their divergence, which is considerably more recent than hitherto assumed, supporting the "recent heritage and fast evolution" scenario for SMP evolution.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Morris SC. The Crucible of Creation: The Burgess Shale and the Rise of Animals. Highlands Ranch: Peterson's; 1998.
-
- Shubin NH, Marshall CR. Fossils, genes, and the origin of novelty. Paleobiology. 2000;26:324–340. doi: 10.1017/S0094837300026993. - DOI
-
- Lowenstam HA, Weiner S. On Biomineralization. Oxford: Oxford University Press on Demand; 1989.
-
- Carter JG, Clark GR. Classification and phylogenetic significance of molluscan shell microstructure. Stud. Geol. Notes Short Course. 1985;13:50–71. doi: 10.1017/S0271164800001093. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

