The global epidemiology of emerging Histoplasma species in recent years
- PMID: 33335607
- PMCID: PMC7714791
- DOI: 10.1016/j.simyco.2020.02.001
The global epidemiology of emerging Histoplasma species in recent years
Abstract
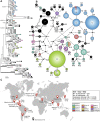
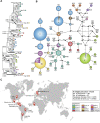
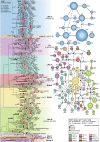
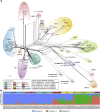
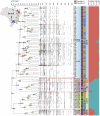
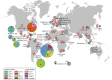
Histoplasmosis is a serious infectious disease in humans caused by Histoplasma spp. (Onygenales), whose natural reservoirs are thought to be soil enriched with bird and bat guano. The true global burden of histoplasmosis is underestimated and frequently the pulmonary manifestations are misdiagnosed as tuberculosis. Molecular data on epidemiology of Histoplasma are still scarce, even though there is increasing recognition of histoplasmosis in recent years in areas distant from the traditional endemic regions in the Americas. We used multi-locus sequence data from protein coding loci (ADP-ribosylation factor, H antigen precursor, and delta-9 fatty acid desaturase), DNA barcoding (ITS1/2+5.8s), AFLP markers and mating type analysis to determine the genetic diversity, population structure and recognise the existence of different phylogenetic species among 436 isolates of Histoplasma obtained globally. Our study describes new phylogenetic species and the molecular characteristics of Histoplasma lineages causing outbreaks with a high number of severe outcomes in Northeast Brazil between 2011 and 2015. Genetic diversity levels provide evidence for recombination, common ancestry and clustering of Brazilian isolates at different geographic scales with the emergence of LAm C, a new genotype assigned to a separate population cluster in Northeast Brazil that exhibited low diversity indicative of isolation. The global survey revealed that the high genetic variability among Brazilian isolates along with the presence of divergent cryptic species and/or genotypes may support the hypothesis of Brazil being the center of dispersion of Histoplasma in South America, possibly with the contribution of migratory hosts such as birds and bats. Outside Brazil, the predominant species depends on the region. We confirm that histoplasmosis has significantly broadened its area of occurrence, an important feature of emerging pathogens. From a practical point of view, our data point to the emergence of histoplasmosis caused by a plethora of genotypes, and will enable epidemiological analysis focused on understanding the processes that lead to histoplasmosis. Further, the description of this diversity opens avenues for comparative genomic studies, which will allow progress toward a consensus taxonomy, improve understanding of the presence of hybrids in natural populations of medically relevant fungi, test reproductive barriers and to explore the significance of this variation.
Keywords: Emerging pathogens; Epidemiology; Genetic diversity; Histoplasma capsulatum; Histoplasmosis; Population structure.
© 2020 Westerdijk Fungal Biodiversity Institute. Production and hosting by ELSEVIER B.V.
Figures


References
-
- Almeida M.A., Almeida-Silva F., Guimaraes A.J. The occurrence of histoplasmosis in Brazil: A systematic review. International Journal of Infectious Diseases. 2019;86:147–156. - PubMed
-
- Altman D.G. Chapman and Hall; London: 1991. Practical statistics for medical research; p. 624.
-
- Ashbee H.R., Evans E.G.V., Viviani M.A. Histoplasmosis in Europe: report on an epidemiological survey from the European Confederation of Medical Mycology Working Group. Medical Mycology. 2008;46:57–65. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
