Linc00475 promotes the progression of glioma by regulating the miR-141-3p/YAP1 axis
- PMID: 33336871
- PMCID: PMC7810941
- DOI: 10.1111/jcmm.16100
Linc00475 promotes the progression of glioma by regulating the miR-141-3p/YAP1 axis
Abstract
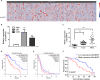
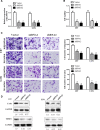
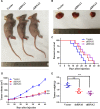
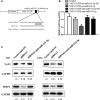
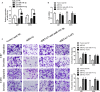
Glioma is the most prevalent and lethal primary brain tumour. Abundant long non-coding RNAs ( lncRNAs) are aberrant and play crucial roles in the oncogenesis of glioma. The exact functions of linc00475 in glioma remain blurred. Here, we analysed the expression levels of linc00475 by qRT-PCR and discovered that linc00475 was up-regulated in glioma and predicted a poor prognosis in patients with glioma. Besides, inhibiting linc00475 restrained the progression of glioma in vitro and in vivo. Further experiments confirmed that linc00475 regulated the progression of glioma by acting as a sponge for miR-141-3p. Moreover, we detected the binding sites of linc00475 and miR-141-3p, the YAP1- 3'UTR and miR-141-3p by luciferase reporters. The rescue assays confirmed that inhibiting linc00475 restrained the progression of glioma through the miR-141-3p/YAP1 pathway. Collectively, our research demonstrates the key roles of linc00475 in glioma, which could be a promising therapeutic target.
Keywords: YAP1; ceRNA; glioma; linc00475; miR-141-3p; prognosis.
© 2020 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Liu B, Cao W, Ma H. Knockdown of lncRNA LSINCT5 suppresses growth and metastasis of human glioma cells via up‐regulating miR‐451. Artif Cells Nanomed Biotechnol. 2019;47:2507‐2515. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

