TMEM41B Is a Pan-flavivirus Host Factor
- PMID: 33338421
- PMCID: PMC7954666
- DOI: 10.1016/j.cell.2020.12.005
TMEM41B Is a Pan-flavivirus Host Factor
Abstract
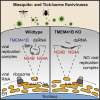
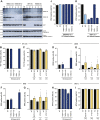
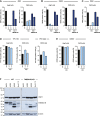
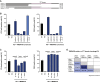
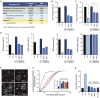
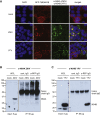
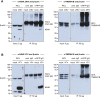
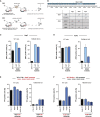
Flaviviruses pose a constant threat to human health. These RNA viruses are transmitted by the bite of infected mosquitoes and ticks and regularly cause outbreaks. To identify host factors required for flavivirus infection, we performed full-genome loss of function CRISPR-Cas9 screens. Based on these results, we focused our efforts on characterizing the roles that TMEM41B and VMP1 play in the virus replication cycle. Our mechanistic studies on TMEM41B revealed that all members of the Flaviviridae family that we tested require TMEM41B. We tested 12 additional virus families and found that SARS-CoV-2 of the Coronaviridae also required TMEM41B for infection. Remarkably, single nucleotide polymorphisms present at nearly 20% in East Asian populations reduce flavivirus infection. Based on our mechanistic studies, we propose that TMEM41B is recruited to flavivirus RNA replication complexes to facilitate membrane curvature, which creates a protected environment for viral genome replication.
Keywords: CRISPR; TMEM41B; VMP1; autophagy; coronavirus; flavivirus; flavivirus replication complex; membrane remodeling.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests C.M. Rice is a founder of Apath LLC; a Scientific Advisory Board member of Imvaq Therapeutics, Vir Biotechnology, and Arbutus Biopharma; and an advisor for Regulus Therapeutics and Pfizer. The remaining authors declare no competing interests. C.M. Rudin serves on the Scientific Advisory Boards of Bridge Medicines, Earli, and Harpoon Therapeutics.
Figures





Update of
-
TMEM41B is a pan-flavivirus host factor.bioRxiv [Preprint]. 2020 Oct 11:2020.10.09.334128. doi: 10.1101/2020.10.09.334128. bioRxiv. 2020. Update in: Cell. 2021 Jan 7;184(1):133-148.e20. doi: 10.1016/j.cell.2020.12.005. PMID: 33052348 Free PMC article. Updated. Preprint.
Comment in
-
A Crisp(r) New Perspective on SARS-CoV-2 Biology.Cell. 2021 Jan 7;184(1):15-17. doi: 10.1016/j.cell.2020.12.003. Epub 2020 Dec 17. Cell. 2021. PMID: 33338422 Free PMC article.
References
-
- Ahmed Q.A., Memish Z.A. Yellow fever from Angola and Congo: a storm gathers. Trop. Doct. 2017;47:92–96. - PubMed
-
- Baggen J., Persoons L., Jansen S., Vanstreels E., Jacquemyn M., Jochmans D., Neyts J., Dallmeier K., Maes P., Daelemans D. Identification of TMEM106B as proviral host factor for SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.09.28.316281. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

