DNA Repair Biosensor-Identified DNA Damage Activities of Endophyte Extracts from Garcinia cowa
- PMID: 33339185
- PMCID: PMC7765599
- DOI: 10.3390/biom10121680
DNA Repair Biosensor-Identified DNA Damage Activities of Endophyte Extracts from Garcinia cowa
Abstract
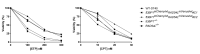
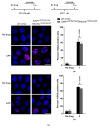
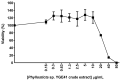
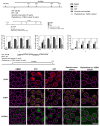
Recent developments in chemotherapy focus on target-specific mechanisms, which occur only in cancer cells and minimize the effects on normal cells. DNA damage and repair pathways are a promising target in the treatment of cancer. In order to identify novel compounds targeting DNA repair pathways, two key proteins, 53BP1 and RAD54L, were tagged with fluorescent proteins as indicators for two major double strand break (DSB) repair pathways: non-homologous end-joining (NHEJ) and homologous recombination (HR). The engineered biosensor cells exhibited the same DNA repair properties as the wild type. The biosensor cells were further used to investigate the DNA repair activities of natural biological compounds. An extract from Phyllosticta sp., the endophyte isolated from the medicinal plant Garcinia cowa Roxb. ex Choisy, was tested. The results showed that the crude extract induced DSB, as demonstrated by the increase in the DNA DSB marker γH2AX. The damaged DNA appeared to be repaired through NHEJ, as the 53BP1 focus formation in the treated fraction was higher than in the control group. In conclusion, DNA repair-based biosensors are useful for the preliminary screening of crude extracts and biological compounds for the identification of potential targeted therapeutic drugs.
Keywords: DNA damage and repair; Garcinia cowa Roxb. ex Choisy; biosensor; cancer; endophyte.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







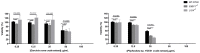
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

