Simultaneous Quantification of Vibrio metoecus and Vibrio cholerae with Its O1 Serogroup and Toxigenic Subpopulations in Environmental Reservoirs
- PMID: 33339261
- PMCID: PMC7766680
- DOI: 10.3390/pathogens9121053
Simultaneous Quantification of Vibrio metoecus and Vibrio cholerae with Its O1 Serogroup and Toxigenic Subpopulations in Environmental Reservoirs
Abstract
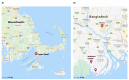
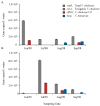
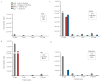
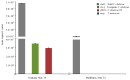
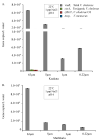
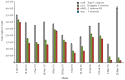
Vibrio metoecus is a recently described aquatic bacterium and opportunistic pathogen, closely related to and often coexisting with Vibrio cholerae. To study the relative abundance and population dynamics of both species in aquatic environments of cholera-endemic and cholera-free regions, we developed a multiplex qPCR assay allowing simultaneous quantification of total V. metoecus and V. cholerae (including toxigenic and O1 serogroup) cells. The presence of V. metoecus was restricted to samples from regions that are not endemic for cholera, where it was found at 20% of the abundance of V. cholerae. In this environment, non-toxigenic O1 serogroup V. cholerae represents almost one-fifth of the total V. cholerae population. In contrast, toxigenic O1 serogroup V. cholerae was also present in low abundance on the coast of cholera-endemic regions, but sustained in relatively high proportions throughout the year in inland waters. The majority of cells from both Vibrio species were recovered from particles rather than free-living, indicating a potential preference for attached versus planktonic lifestyles. This research further elucidates the population dynamics underpinning V. cholerae and its closest relative in cholera-endemic and non-endemic regions through culture-independent quantification from environmental samples.
Keywords: O1; Vibrio cholerae; Vibrio metoecus; cholera-endemic; qPCR; serogroup; toxigenic and non-toxigenic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Alam M., Hasan N.A., Sadique A., Bhuiyan N.A., Ahmed K.U., Nusrin S., Nair G.B., Siddique A.K., Sack R.B., Sack D.A., et al. Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl. Environ. Microbiol. 2006;72:4096–4104. doi: 10.1128/AEM.00066-06. - DOI - PMC - PubMed
-
- WHO Number of Reported Cholera Cases. [(accessed on 27 August 2020)]; Available online: https://www.who.int/gho/epidemic_diseases/cholera/cases_text/en/
Grants and funding
LinkOut - more resources
Full Text Sources

