The Potential of Proteolytic Chimeras as Pharmacological Tools and Therapeutic Agents
- PMID: 33339292
- PMCID: PMC7766482
- DOI: 10.3390/molecules25245956
The Potential of Proteolytic Chimeras as Pharmacological Tools and Therapeutic Agents
Abstract
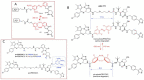
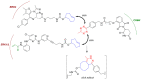
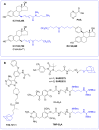
The induction of protein degradation in a highly selective and efficient way by means of druggable molecules is known as targeted protein degradation (TPD). TPD emerged in the literature as a revolutionary idea: a heterobifunctional chimera with the capacity of creating an interaction between a protein of interest (POI) and a E3 ubiquitin ligase will induce a process of events in the POI, including ubiquitination, targeting to the proteasome, proteolysis and functional silencing, acting as a sort of degradative knockdown. With this programmed protein degradation, toxic and disease-causing proteins could be depleted from cells with potentially effective low drug doses. The proof-of-principle validation of this hypothesis in many studies has made the TPD strategy become a new attractive paradigm for the development of therapies for the treatment of multiple unmet diseases. Indeed, since the initial protacs (Proteolysis targeting chimeras) were posited in the 2000s, the TPD field has expanded extraordinarily, developing innovative chemistry and exploiting multiple degradation approaches. In this article, we review the breakthroughs and recent novel concepts in this highly active discipline.
Keywords: autophagy; chimera; lysosome; protac; proteasome; targeted protein degradation; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Advances in designing ternary complexes: Integrating in-silico and biochemical methods for PROTAC optimisation in target protein degradation.Bioorg Chem. 2024 Dec;153:107868. doi: 10.1016/j.bioorg.2024.107868. Epub 2024 Oct 4. Bioorg Chem. 2024. PMID: 39374557 Review.
-
Small-Molecule Degraders beyond PROTACs-Challenges and Opportunities.SLAS Discov. 2021 Apr;26(4):524-533. doi: 10.1177/2472555221991104. Epub 2021 Feb 25. SLAS Discov. 2021. PMID: 33632029 Review.
-
Perspectives on the development of first-in-class protein degraders.Future Med Chem. 2021 Jul;13(14):1203-1226. doi: 10.4155/fmc-2021-0033. Epub 2021 May 21. Future Med Chem. 2021. PMID: 34015962 Review.
-
PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics.Oncogene. 2020 Jun;39(26):4909-4924. doi: 10.1038/s41388-020-1336-y. Epub 2020 May 31. Oncogene. 2020. PMID: 32475992 Free PMC article. Review.
-
Ligandability of E3 Ligases for Targeted Protein Degradation Applications.Biochemistry. 2023 Feb 7;62(3):588-600. doi: 10.1021/acs.biochem.1c00464. Epub 2021 Sep 2. Biochemistry. 2023. PMID: 34473924 Free PMC article. Review.
Cited by
-
The proteolysis targeting chimera GMB-475 combined with dasatinib for the treatment of chronic myeloid leukemia with BCR::ABL1 mutants.Front Pharmacol. 2022 Oct 3;13:931772. doi: 10.3389/fphar.2022.931772. eCollection 2022. Front Pharmacol. 2022. PMID: 36263131 Free PMC article.
-
Designing Bioorthogonal Reactions for Biomedical Applications.Research (Wash D C). 2023 Dec 15;6:0251. doi: 10.34133/research.0251. eCollection 2023. Research (Wash D C). 2023. PMID: 38107023 Free PMC article. Review.
-
Opportunities and challenges in translational science.Clin Transl Sci. 2021 Sep;14(5):1629-1647. doi: 10.1111/cts.13055. Epub 2021 Jul 8. Clin Transl Sci. 2021. PMID: 33982407 Free PMC article. Review.
-
Protein aggregation and therapeutic strategies in SOD1- and TDP-43- linked ALS.Front Mol Biosci. 2024 May 24;11:1383453. doi: 10.3389/fmolb.2024.1383453. eCollection 2024. Front Mol Biosci. 2024. PMID: 38855322 Free PMC article. Review.
-
Direct targeting of TDP-43, from small molecules to biologics: the therapeutic landscape.RSC Chem Biol. 2021 Jun 21;2(4):1158-1166. doi: 10.1039/d1cb00110h. eCollection 2021 Aug 5. RSC Chem Biol. 2021. PMID: 34458829 Free PMC article. Review.

