In Vivo Evidence for Serine Biosynthesis-Defined Sensitivity of Lung Metastasis, but Not of Primary Breast Tumors, to mTORC1 Inhibition
- PMID: 33340488
- PMCID: PMC9161668
- DOI: 10.1016/j.molcel.2020.11.027
In Vivo Evidence for Serine Biosynthesis-Defined Sensitivity of Lung Metastasis, but Not of Primary Breast Tumors, to mTORC1 Inhibition
Abstract
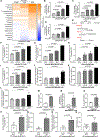
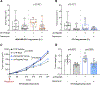
In tumors, nutrient availability and metabolism are known to be important modulators of growth signaling. However, it remains elusive whether cancer cells that are growing out in the metastatic niche rely on the same nutrients and metabolic pathways to activate growth signaling as cancer cells within the primary tumor. We discovered that breast-cancer-derived lung metastases, but not the corresponding primary breast tumors, use the serine biosynthesis pathway to support mTORC1 growth signaling. Mechanistically, pyruvate uptake through Mct2 supported mTORC1 signaling by fueling serine biosynthesis-derived α-ketoglutarate production in breast-cancer-derived lung metastases. Consequently, expression of the serine biosynthesis enzyme PHGDH was required for sensitivity to the mTORC1 inhibitor rapamycin in breast-cancer-derived lung tumors, but not in primary breast tumors. In summary, we provide in vivo evidence that the metabolic and nutrient requirements to activate growth signaling differ between the lung metastatic niche and the primary breast cancer site.
Keywords: MCT2; PHGDH; breast cancer; lung environment; mTORC1; metastasis formation; pyruvate; serine biosynthesis; α-ketoglutarate.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests S.-M.F. has received funding from Bayer AG, Merck, and Black Belt Therapeutics; has consulted for Fund+; and serves on the advisory board of Molecular Cell. All other authors declare no competing interests.
Figures



Comment in
-
Food for Growth: Distinct Nutrient Preferences between Primary Tumors and Metastases.Mol Cell. 2021 Jan 21;81(2):220-222. doi: 10.1016/j.molcel.2020.12.044. Mol Cell. 2021. PMID: 33482090
References
-
- Altea-Manzano P, Broekaert D, Duarte JAG, Ferná ndez-García J, Planque M, and Fendt S-M (2020). Analyzing the Metabolism of Metastases in Mice. Methods Mol. Biol. 2088, 93–118. - PubMed
-
- Bligh EG, and Dyer WJ (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

