Single-Cell Transcriptomics Reveals Zone-Specific Alterations of Liver Sinusoidal Endothelial Cells in Cirrhosis
- PMID: 33340713
- PMCID: PMC7903131
- DOI: 10.1016/j.jcmgh.2020.12.007
Single-Cell Transcriptomics Reveals Zone-Specific Alterations of Liver Sinusoidal Endothelial Cells in Cirrhosis
Abstract
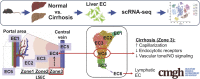
Background: Dysfunction of liver sinusoidal endothelial cells (LSECs) is permissive for the progression of liver fibrosis and cirrhosis and responsible for its clinical complications. Here, we have mapped the spatial distribution of heterogeneous liver ECs in normal vs cirrhotic mouse livers and identified zone-specific transcriptomic changes of LSECs associated with liver cirrhosis using scRNA-seq technology.
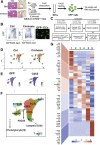
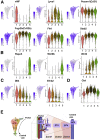
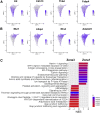
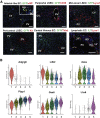
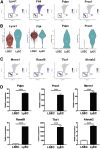
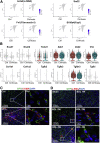
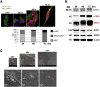
Approach & results: Cirrhosis was generated in endothelial specific green fluorescent protein (GFP) reporter mice through carbon tetrachloride inhalation for 12 weeks. GFP-positive liver EC populations were isolated from control and cirrhotic mice by FACS. We identified 6 clusters of liver EC populations including 3 clusters of LSECs, 2 clusters of vascular ECs and 1 cluster of lymphatic ECs. Based on previously reported LSEC-landmarks, we mapped the 3 clusters of LSECs in zones 1, 2, and 3, and determined phenotypic changes in each zone between control and cirrhotic mice. We found genes representing capillarization of LSECs (eg, CD34) as well as extracellular matrix genes were most upregulated in LSECs of zone 3 in cirrhotic mice, which may contribute to the development of basement membranes. LSECs in cirrhotic mice also demonstrated decreased expression of endocytic receptors, most remarkably in zone 3. Transcription factors (Klf2 [Kruppel-like factor-2], Klf4 [Kruppel-like factor-4], and AP-1) that induce nitric oxide production in response to shear stress were downregulated in LSECs of all zones in cirrhotic mice, implying increased intrahepatic vascular resistance.
Conclusion: This study deepens our knowledge of the pathogenesis of liver cirrhosis at a spatial, cell-specific level, which is indispensable for the development of novel therapeutic strategies to target the most dysfunctional liver ECs.
Keywords: Endothelial Dysfunction; Liver Fibrosis; Lymphatic Endothelial Cells; Portal Hypertension; scRNA-seq.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Halpern K.B., Shenhav R., Matcovitch-Natan O., Toth B., Lemze D., Golan M., Massasa E.E., Baydatch S., Landen S., Moor A.E., Brandis A., Giladi A., Avihail A.S., David E., Amit I., Itzkovitz S. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. - PMC - PubMed
-
- Dobie R., Wilson-Kanamori J.R., Henderson B.E.P., Smith J.R., Matchett K.P., Portman J.R., Wallenborg K., Picelli S., Zagorska A., Pendem S.V., Hudson T.E., Wu M.M., Budas G.R., Breckenridge D.G., Harrison E.M., Mole D.J., Wigmore S.J., Ramachandran P., Ponting C.P., Teichmann S.A., Marioni J.C., Henderson N.C. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep. 2019;29:1832–1847.e8. - PMC - PubMed
-
- MacParland S.A., Liu J.C., Ma X.Z., Innes B.T., Bartczak A.M., Gage B.K., Manuel J., Khuu N., Echeverri J., Linares I., Gupta R., Cheng M.L., Liu L.Y., Camat D., Chung S.W., Seliga R.K., Shao Z., Lee E., Ogawa S., Ogawa M., Wilson M.D., Fish J.E., Selzner M., Ghanekar A., Grant D., Greig P., Sapisochin G., Selzner N., Winegarden N., Adeyi O., Keller G., Bader G.D., McGilvray I.D. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

