Heterologous expression of the atypical tetracycline chelocardin reveals the full set of genes required for its biosynthesis
- PMID: 33341113
- PMCID: PMC7749508
- DOI: 10.1186/s12934-020-01495-x
Heterologous expression of the atypical tetracycline chelocardin reveals the full set of genes required for its biosynthesis
Abstract
Background: Chelocardin (CHD) exhibits a broad-spectrum antibiotic activity and showed promising results in a small phase II clinical study conducted on patients with urinary tract infections. Importantly, CHD was shown to be active also against tetracycline-resistant Gram-negative pathogens, which is gaining even more importance in today's antibiotic crisis. We have demonstrated that modifications of CHD through genetic engineering of its producer, the actinomycete Amycolatopsis sulphurea, are not only possible but yielded even more potent antibiotics than CHD itself, like 2-carboxamido-2-deacetyl-chelocardin (CD-CHD), which is currently in preclinical evaluation. A. sulphurea is difficult to genetically manipulate and therefore manipulation of the chd biosynthetic gene cluster in a genetically amenable heterologous host would be of high importance for further drug-discovery efforts.
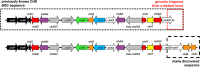
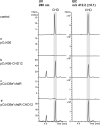
Results: We report heterologous expression of the CHD biosynthetic gene cluster in the model organism Streptomyces albus del14 strain. Unexpectedly, we found that the originally defined CHD gene cluster fails to provide all genes required for CHD formation, including an additional cyclase and two regulatory genes. Overexpression of the putative pathway-specific streptomyces antibiotic regulatory protein chdB in A. sulphurea resulted in an increase of both, CHD and CD-CHD production. Applying a metabolic-engineering approach, it was also possible to generate the potent CHD analogue, CD-CHD in S. albus. Finally, an additional yield increase was achieved in S. albus del14 by in-trans overexpression of the chdR exporter gene, which provides resistance to CHD and CDCHD.
Conclusions: We identified previously unknown genes in the CHD cluster, which were shown to be essential for chelocardin biosynthesis by expression of the full biosynthetic gene cluster in S. albus as heterologous host. When comparing to oxytetracycline biosynthesis, we observed that the CHD gene cluster contains additional enzymes not found in gene clusters encoding the biosynthesis of typical tetracyclines (such as oxytetracycline). This finding probably explains the different chemistries and modes of action, which make CHD/CD-CHD valuable lead structures for clinical candidates. Even though the CHD genes are derived from a rare actinomycete A. sulphurea, the yield of CHD in the heterologous host was very good. The corrected nucleotide sequence of the CHD gene cluster now contains all gene products required for the production of CHD in a genetically amenable heterologous host, thus opening new possibilities towards production of novel and potent tetracycline analogues with a new mode of action.
Keywords: Actinobacteria; Antibiotics; Chelocardin; Heterologous expression; Natural product biosynthesis; Polyketide; Tetracyclines.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
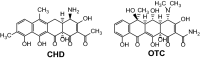



Similar articles
-
Engineering Atypical Tetracycline Formation in Amycolatopsis sulphurea for the Production of Modified Chelocardin Antibiotics.ACS Chem Biol. 2019 Mar 15;14(3):468-477. doi: 10.1021/acschembio.8b01125. Epub 2019 Feb 12. ACS Chem Biol. 2019. PMID: 30747520
-
Identification of the chelocardin biosynthetic gene cluster from Amycolatopsis sulphurea: a platform for producing novel tetracycline antibiotics.Microbiology (Reading). 2013 Dec;159(Pt 12):2524-2532. doi: 10.1099/mic.0.070995-0. Epub 2013 Sep 16. Microbiology (Reading). 2013. PMID: 24043447
-
The ADEP Biosynthetic Gene Cluster in Streptomyces hawaiiensis NRRL 15010 Reveals an Accessory clpP Gene as a Novel Antibiotic Resistance Factor.Appl Environ Microbiol. 2019 Oct 1;85(20):e01292-19. doi: 10.1128/AEM.01292-19. Print 2019 Oct 15. Appl Environ Microbiol. 2019. PMID: 31399403 Free PMC article.
-
Streptomycetes: Surrogate hosts for the genetic manipulation of biosynthetic gene clusters and production of natural products.Biotechnol Adv. 2019 Jan-Feb;37(1):1-20. doi: 10.1016/j.biotechadv.2018.10.003. Epub 2018 Oct 9. Biotechnol Adv. 2019. PMID: 30312648 Free PMC article. Review.
-
Challenges in the Heterologous Production of Antibiotics in Streptomyces.Arch Pharm (Weinheim). 2016 Aug;349(8):594-601. doi: 10.1002/ardp.201600058. Epub 2016 Jun 3. Arch Pharm (Weinheim). 2016. PMID: 27258165 Review.
Cited by
-
Pharmacokinetic and pharmacodynamic evaluation of the atypical tetracyclines chelocardin and amidochelocardin in murine infection models.Microbiol Spectr. 2024 Jan 11;12(1):e0128923. doi: 10.1128/spectrum.01289-23. Epub 2023 Dec 4. Microbiol Spectr. 2024. PMID: 38047701 Free PMC article.
-
Looking Back to Amycolatopsis: History of the Antibiotic Discovery and Future Prospects.Antibiotics (Basel). 2021 Oct 15;10(10):1254. doi: 10.3390/antibiotics10101254. Antibiotics (Basel). 2021. PMID: 34680834 Free PMC article. Review.
-
An Improved Transformation-Associated Recombination Cloning Approach for Direct Capturing of Natural Product Biosynthetic Gene Clusters.Microb Biotechnol. 2024 Dec;17(12):e70067. doi: 10.1111/1751-7915.70067. Microb Biotechnol. 2024. PMID: 39651843 Free PMC article.
References
-
- Lukezic T, Lesnik U, Podgorsek A, Horvat J, Polak T, Sala M, et al. Identification of the chelocardin biosynthetic gene cluster from Amycolatopsis sulphurea: a platform for producing novel tetracycline antibiotics. Microbiology. 2013;159(Pt 12):2524–2532. - PubMed
-
- Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat Prod Rep. 2007;24(1):162–190. - PubMed
-
- Stepanek JJ, Lukežič T, Teichert I, Petković H, Bandow JE. Dual mechanism of action of the atypical tetracycline chelocardin. Biochim Biophys Acta. 2016;1864(6):645–654. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

