Simulation of African and non-African low and high coverage whole genome sequence data to assess variant calling approaches
- PMID: 33341897
- PMCID: PMC8294538
- DOI: 10.1093/bib/bbaa366
Simulation of African and non-African low and high coverage whole genome sequence data to assess variant calling approaches
Abstract
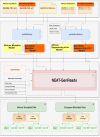
Current variant calling (VC) approaches have been designed to leverage populations of long-range haplotypes and were benchmarked using populations of European descent, whereas most genetic diversity is found in non-European such as Africa populations. Working with these genetically diverse populations, VC tools may produce false positive and false negative results, which may produce misleading conclusions in prioritization of mutations, clinical relevancy and actionability of genes. The most prominent question is which tool or pipeline has a high rate of sensitivity and precision when analysing African data with either low or high sequence coverage, given the high genetic diversity and heterogeneity of this data. Here, a total of 100 synthetic Whole Genome Sequencing (WGS) samples, mimicking the genetics profile of African and European subjects for different specific coverage levels (high/low), have been generated to assess the performance of nine different VC tools on these contrasting datasets. The performances of these tools were assessed in false positive and false negative call rates by comparing the simulated golden variants to the variants identified by each VC tool. Combining our results on sensitivity and positive predictive value (PPV), VarDict [PPV = 0.999 and Matthews correlation coefficient (MCC) = 0.832] and BCFtools (PPV = 0.999 and MCC = 0.813) perform best when using African population data on high and low coverage data. Overall, current VC tools produce high false positive and false negative rates when analysing African compared with European data. This highlights the need for development of VC approaches with high sensitivity and precision tailored for populations characterized by high genetic variations and low linkage disequilibrium.
Keywords: DNA sequence; genomics; next-generation sequence; simulation; variant calling.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

