A Novel Text-Mining Approach for Retrieving Pharmacogenomics Associations From the Literature
- PMID: 33343371
- PMCID: PMC7748107
- DOI: 10.3389/fphar.2020.602030
A Novel Text-Mining Approach for Retrieving Pharmacogenomics Associations From the Literature
Abstract
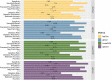
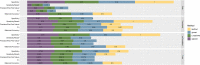
Text mining in biomedical literature is an emerging field which has already been shown to have a variety of implementations in many research areas, including genetics, personalized medicine, and pharmacogenomics. In this study, we describe a novel text-mining approach for the extraction of pharmacogenomics associations. The code that was used toward this end was implemented using R programming language, either through custom scripts, where needed, or through utilizing functions from existing libraries. Articles (abstracts or full texts) that correspond to a specified query were extracted from PubMed, while concept annotations were derived by PubTator Central. Terms that denote a Mutation or a Gene as well as Chemical compound terms corresponding to drug compounds were normalized and the sentences containing the aforementioned terms were filtered and preprocessed to create appropriate training sets. Finally, after training and adequate hyperparameter tuning, four text classifiers were created and evaluated (FastText, Linear kernel SVMs, XGBoost, Lasso, and Elastic-Net Regularized Generalized Linear Models) with regard to their performance in identifying pharmacogenomics associations. Although further improvements are essential toward proper implementation of this text-mining approach in the clinical practice, our study stands as a comprehensive, simplified, and up-to-date approach for the identification and assessment of research articles enriched in clinically relevant pharmacogenomics relationships. Furthermore, this work highlights a series of challenges concerning the effective application of text mining in biomedical literature, whose resolution could substantially contribute to the further development of this field.
Keywords: FastText, biomedical text classification, supervised learning; Pubmed; Pubtator; natural language processing; pharmacogenomics associations; text mining.
Copyright © 2020 Patrinos, Pandi, Van Der Spek and Koromina.
Figures

References
-
- Benesty M. (2019). Fastrtext: ‘fastText’ wrapper for text classification and word representation. R Foundation for Statistical Computing.
-
- Chen T., Guestrin C. (2016). “XGBoost: a scalable tree boosting system,” in Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining (San Francisco, CA: Association for Computing Machinery; ), 785–794.
-
- Dmitriy Selivanov M. B., Wang Q. (2020). text2vec: modern text mining framework for R. R Foundation for Statistical Computing.
LinkOut - more resources
Full Text Sources

