Induction of the cydAB Operon Encoding the bd Quinol Oxidase Under Respiration-Inhibitory Conditions by the Major cAMP Receptor Protein MSMEG_6189 in Mycobacterium smegmatis
- PMID: 33343552
- PMCID: PMC7739888
- DOI: 10.3389/fmicb.2020.608624
Induction of the cydAB Operon Encoding the bd Quinol Oxidase Under Respiration-Inhibitory Conditions by the Major cAMP Receptor Protein MSMEG_6189 in Mycobacterium smegmatis
Abstract
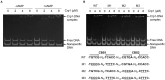
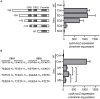
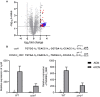
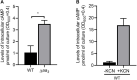
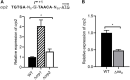
The respiratory electron transport chain (ETC) of Mycobacterium smegmatis is terminated with two terminal oxidases, the aa 3 cytochrome c oxidase and the cytochrome bd quinol oxidase. The bd quinol oxidase with a higher binding affinity for O2 than the aa 3 oxidase is known to play an important role in aerobic respiration under oxygen-limiting conditions. Using relevant crp1 (MSMEG_6189) and crp2 (MSMEG_0539) mutant strains of M. smegmatis, we demonstrated that Crp1 plays a predominant role in induction of the cydAB operon under ETC-inhibitory conditions. Two Crp-binding sequences were identified upstream of the cydA gene, both of which are necessary for induction of cydAB expression under ETC-inhibitory conditions. The intracellular level of cAMP in M. smegmatis was found to be increased under ETC-inhibitory conditions. The crp2 gene was found to be negatively regulated by Crp1 and Crp2, which appears to lead to significantly low cellular abundance of Crp2 relative to Crp1 in M. smegmatis. Our RNA sequencing analyses suggest that in addition to the SigF partner switching system, Crp1 is involved in induction of gene expression in M. smegmatis exposed to ETC-inhibitory conditions.
Keywords: Crp; Mycobacterium; aa3 cytochrome c oxidase; cAMP; electron transport chain; regulation of gene expression; respiration.
Copyright © 2020 Ko and Oh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

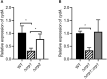


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

