A Streamlined Approach to Rapidly Detect SARS-CoV-2 Infection Avoiding RNA Extraction: Workflow Validation
- PMID: 33343767
- PMCID: PMC7727018
- DOI: 10.1155/2020/8869424
A Streamlined Approach to Rapidly Detect SARS-CoV-2 Infection Avoiding RNA Extraction: Workflow Validation
Abstract
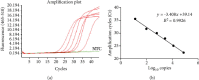
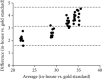
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has rapidly spread worldwide from the beginning of 2020. The presence of viral RNA in samples by nucleic acid (NA) molecular analysis is the only method available to diagnose COVID-19 disease and to assess patients' viral load. Since the demand for laboratory reagents has increased, there has been a worldwide shortage of RNA extraction kits. We, therefore, developed a fast and cost-effective viral genome isolation method that, combined with quantitative RT-PCR assay, detects SARS-CoV-2 RNA in patient samples. The method relies on the addition of Proteinase K followed by a controlled heat-shock incubation and, then, E gene evaluation by RT-qPCR. It was validated for sensitivity, specificity, linearity, reproducibility, and precision. It detects as low as 10 viral copies/sample, is rapid, and has been characterized in 60 COVID-19-infected patients. Compared to automated extraction methods, our pretreatment guarantees the same positivity rate with the advantage of shortening the time of the analysis and reducing its cost. This is a rapid workflow meant to aid the healthcare system in the rapid identification of infected patients, such as during a pathogen-related outbreak. For its intrinsic characteristics, this workflow is suitable for large-scale screenings.
Copyright © 2020 Catia Mio et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Rabaan A. A., Al-Ahmed S. H., Haque S., et al. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Le Infezioni in Medicina. 2020;28(2):174–184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

