Genomic epidemiology of coxsackievirus A16 in mainland of China, 2000-18
- PMID: 33343924
- PMCID: PMC7733612
- DOI: 10.1093/ve/veaa084
Genomic epidemiology of coxsackievirus A16 in mainland of China, 2000-18
Abstract
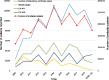
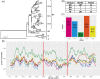
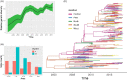
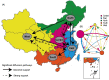
Hand, foot, and mouth disease (HFMD), which is a frequently reported and concerning disease worldwide, is a severe burden on societies globally, especially in the countries of East and Southeast Asia. Coxsackievirus A16 (CV-A16) is one of the most important causes of HFMD and a severe threat to human health, especially in children under 5 years of age. To investigate the epidemiological characteristics, spread dynamics, recombinant forms (RFs), and other features of CV-A16, we leveraged the continuous surveillance data of CV-A16-related HFMD cases collected over an 18-year period. With the advent of the EV-A71 vaccine since 2016, which targeted the EV-A71-related HFMD cases, EV-A71-related HFMD cases decreased dramatically, whereas the CV-A16-related HFMD cases showed an upward trend from 2017 to October 2019. The CV-A16 strains observed in this study were genetically related and widely distributed in the mainland of China. Our results show that three clusters (B1a-B1c) existed in the mainland of China and that the cluster of B1b dominates the diffusion of CV-A16 in China. We found that eastern China played a decisive role in seeding the diffusion of CV-A16 in China, with a more complex and variant transmission trend. Although EV-A71 vaccine was launched in China in 2016, it did not affect the genetic diversity of CV-A16, and its genetic diversity did not decline, which confirmed the epidemiological surveillance trend of CV-A16. Two discontinuous clusters (2000-13 and 2014-18) were observed in the full-length genome and arranged along the time gradient, which revealed the reason why the relative genetic diversity of CV-A16 increased and experienced more complex fluctuation model after 2014. In addition, the switch from RFs B (RF-B) and RF-C co-circulation to RF-D contributes to the prevalence of B1b cluster in China after 2008. The correlation between genotype and RFs partially explained the current prevalence of B1b. This study provides unprecedented full-length genomic sequences of CV-A16 in China, with a wider geographic distribution and a long-term time scale. The study presents valuable information about CV-A16, aimed at developing effective control strategies, as well as a call for a more robust surveillance system, especially in the Asia-Pacific region.
Keywords: coxsackievirus A16; enterovirus; epidemiology; phylodynamics; phylogeny.
© The Author(s) 2020. Published by Oxford University Press.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

