Genomic and Phenotypic Divergence in Wild Barley Driven by Microgeographic Adaptation
- PMID: 33344112
- PMCID: PMC7740101
- DOI: 10.1002/advs.202000709
Genomic and Phenotypic Divergence in Wild Barley Driven by Microgeographic Adaptation
Abstract
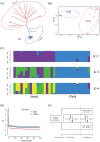
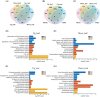
Microgeographic adaptation is a fundamental driving force of evolution, but the underlying causes remain undetermined. Here, the phenotypic, genomic and transcriptomic variations of two wild barley populations collected from sharply divergent and adjacent micro-geographic sites to identify candidate genes associated with edaphic local adaptation are investigated. Common garden and reciprocal transplant studies show that large phenotypic differentiation and local adaptation to soils occur between these populations. Genetic, phylogenetic and admixture analyses based on population resequencing show that significant genetic divergences occur between basalt and chalk populations. These divergences are consistent with the phenotypic variations observed in the field. Genome sweep analyses reveal 162.7 Mb of selected regions driven by edaphic local adaptation, in which 445 genes identified, including genes associated with root architecture, metal transport/detoxification, and ABA signaling. When the phenotypic, genomic and transcriptomic data are combined, HvMOR, encoding an LBD transcription factor, is determined to be the vital candidate for regulating the root architecture to adapt to edaphic conditions at the microgeographic scale. This study provides new insights into the genetic basis of edaphic adaptation and demonstrates that edaphic factors may contribute to the evolution and speciation of barley.
Keywords: adaptive evolution; edaphic adaptation; genetic diversity; whole genome resequencing; wild barley.
© 2020 The Authors. Published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Richardson J. R., Urban M. C., Evolution 2013, 67, 1729. - PubMed
-
- Kawecki T. J., Ebert D., Ecol. Lett. 2004, 7, 1225.
-
- Schluter D., Science 2009, 323, 737. - PubMed
-
- Rajakaruna N., Int. Geol. Rev. 2008, 46, 471.
-
- Richardson J. L., Urban M. C., Bolnick D. I., Skelly D. K., Trends Ecol. Evol. 2014, 29, 165. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
