Emergence and Phylogenetic Analysis of a Getah Virus Isolated in Southern China
- PMID: 33344520
- PMCID: PMC7744783
- DOI: 10.3389/fvets.2020.552517
Emergence and Phylogenetic Analysis of a Getah Virus Isolated in Southern China
Abstract
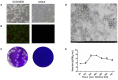
Getah virus (GETV) has caused many outbreaks in animals in recent years. Monitoring of the virus and its related diseases is crucial to control the transmission of the virus. In the summer of 2018, we conducted routine tests on clinical samples from different pig farms in Guangxi province, South China, and isolated and characterized a GETV strain, named GX201808. Cytopathic effects were observed in BHK-21 cells inoculated with GX201808. The expression of E2 protein of GETV could be detected in virus-infected cells by indirect immunofluorescence assays. Electron microscopic analysis showed that the virus particles were spherical and ~70 nm in diameter with featured surface fibers. The multistep growth curves showed the virus propagated well in the BHK-21 cells. Molecular genetic analysis revealed that GX201808 belongs to Group 3, represented by Kochi-01-2005 isolated in Japan in 2005, and it clustered closely with the recently reported Chinese strains isolated from pigs, cattle, and foxes. A comparison of the identities of nucleotides and amino acids in the coding regions demonstrated that the GX201808 showed the highest amino acid identity (99.6%) with the HuN1 strain, a highly pathogenic isolate resulting in an outbreak of GETV infection in swine herds in Hunan province in 2017. In the present study, GETV was identified and isolated for the first time in Guangxi province of southern China, suggesting that future surveillance of this virus should be strengthened.
Keywords: Getah virus; emergence; genetic analysis; isolation; phylogenetic analysis.
Copyright © 2020 Ren, Mo, Wang, Wang, Nong, Wang, Niu, Liu, Chen, Ouyang, Huang and Wei.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Getah Virus (Alphavirus): An Emerging, Spreading Zoonotic Virus.Pathogens. 2022 Aug 20;11(8):945. doi: 10.3390/pathogens11080945. Pathogens. 2022. PMID: 36015065 Free PMC article. Review.
-
First isolation and characterization of Getah virus from cattle in northeastern China.BMC Vet Res. 2019 Sep 5;15(1):320. doi: 10.1186/s12917-019-2061-z. BMC Vet Res. 2019. PMID: 31488162 Free PMC article.
-
Isolation and Characterization of a Porcine Getah Virus Strain from Sichuan Province.Vet Sci. 2025 Mar 15;12(3):276. doi: 10.3390/vetsci12030276. Vet Sci. 2025. PMID: 40267012 Free PMC article.
-
Seroepidemiological investigation of Getah virus in the China-Myanmar border area from 2022-2023.Front Microbiol. 2023 Dec 14;14:1309650. doi: 10.3389/fmicb.2023.1309650. eCollection 2023. Front Microbiol. 2023. PMID: 38163077 Free PMC article.
-
Getah virus as an equine pathogen.Vet Clin North Am Equine Pract. 2000 Dec;16(3):605-17. doi: 10.1016/s0749-0739(17)30099-8. Vet Clin North Am Equine Pract. 2000. PMID: 11219353 Review.
Cited by
-
Outbreak and epidemic of Getah virus infection in swine by virulence-enhanced GIII variant in Henan, central China in 2024.Virulence. 2025 Dec;16(1):2530661. doi: 10.1080/21505594.2025.2530661. Epub 2025 Jul 13. Virulence. 2025. PMID: 40653751 Free PMC article.
-
Early Genomic Surveillance and Phylogeographic Analysis of Getah Virus, a Reemerging Arbovirus, in Livestock in China.J Virol. 2023 Jan 31;97(1):e0109122. doi: 10.1128/jvi.01091-22. Epub 2022 Dec 8. J Virol. 2023. PMID: 36475767 Free PMC article.
-
Rapid Differential Detection of Japanese Encephalitis Virus and Getah Virus in Pigs or Mosquitos by a Duplex TaqMan Real-Time RT-PCR Assay.Front Vet Sci. 2022 Apr 7;9:839443. doi: 10.3389/fvets.2022.839443. eCollection 2022. Front Vet Sci. 2022. PMID: 35464361 Free PMC article.
-
Evolutionary characterization and pathogenicity of Getah virus from pigs in Guangdong Province of China.Arch Virol. 2023 Sep 28;168(10):258. doi: 10.1007/s00705-023-05886-4. Arch Virol. 2023. PMID: 37770803
-
Getah Virus (Alphavirus): An Emerging, Spreading Zoonotic Virus.Pathogens. 2022 Aug 20;11(8):945. doi: 10.3390/pathogens11080945. Pathogens. 2022. PMID: 36015065 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

