Phosphoproteomics to Characterize Host Response During H3N2 Canine Influenza Virus Infection of Dog Lung
- PMID: 33344528
- PMCID: PMC7744373
- DOI: 10.3389/fvets.2020.585071
Phosphoproteomics to Characterize Host Response During H3N2 Canine Influenza Virus Infection of Dog Lung
Abstract
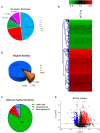
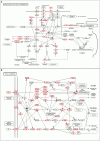
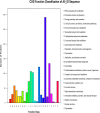
Avian-origin H3N2 canine influenza viruses (CIVs) cause severe contagious respiratory disease in dogs, and quickly adapt to new environments. To further understand the mechanism of virus infection and host-virus interactions, we characterized the complete phosphoproteome of dogs infected with H3N2 CIV. Nine-week-old Beagle dogs were inoculated intranasally with 106 EID50 of A/canine/Guangdong/04/2014 (H3N2) virus. Lung sections were harvested at 5 days post-inoculation (dpi) and processed for global and quantitative analysis of differentially expressed phosphoproteins. A total of 1,235 differentially expressed phosphorylated proteins were identified in the dog lung after H3N2 CIV infection, and 3,016 modification sites were identified among all differentially expressed proteins. We then performed an enrichment analysis of functional annotations using Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology (GO) database analyses to predict the functions of the identified differential phosphoproteins. Our data indicate that H3N2 CIV infection causes dramatic changes in the host protein phosphorylation of dog lungs. To our knowledge, this is the first study to assess the effect of H3N2 CIV infection on the phosphoproteome of beagles. These data provide novel insights into H3N2-CIV-triggered regulatory phosphorylation circuits and signaling networks and may improve our understanding of the mechanisms underlying CIV pathogenesis in dogs.
Keywords: H3N2; KEGG; canine influenza; dog; go; phosphoproteomics.
Copyright © 2020 Liu, Fu, Ye, Liang, Qi, Yao, Wang, Wang, Cai, Tang, Chen and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Transmission and pathogenicity of canine H3N2 influenza virus in dog and guinea pig models.Virol J. 2022 Oct 12;19(1):162. doi: 10.1186/s12985-022-01888-x. Virol J. 2022. PMID: 36224594 Free PMC article.
-
Comparative pathogenesis of H3N2 canine influenza virus in beagle dogs challenged by intranasal and intratracheal inoculation.Virus Res. 2018 Aug 15;255:147-153. doi: 10.1016/j.virusres.2018.05.023. Epub 2018 May 31. Virus Res. 2018. PMID: 29860092
-
Zoonotic Risk, Pathogenesis, and Transmission of Avian-Origin H3N2 Canine Influenza Virus.J Virol. 2017 Oct 13;91(21):e00637-17. doi: 10.1128/JVI.00637-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28814512 Free PMC article.
-
Avian-origin H3N2 canine influenza virus circulating in farmed dogs in Guangdong, China.Infect Genet Evol. 2013 Mar;14:444-9. doi: 10.1016/j.meegid.2012.11.018. Epub 2012 Dec 20. Infect Genet Evol. 2013. Corrected and republished in: Infect Genet Evol. 2013 Oct;19:251-6. doi: 10.1016/j.meegid.2013.05.022. PMID: 23261544 Corrected and republished.
-
H3N8 and H3N2 Canine Influenza Viruses: Understanding These New Viruses in Dogs.Vet Clin North Am Small Anim Pract. 2019 Jul;49(4):643-649. doi: 10.1016/j.cvsm.2019.02.005. Epub 2019 Apr 4. Vet Clin North Am Small Anim Pract. 2019. PMID: 30956002 Review.
Cited by
-
Knowledge, Attitudes, and Risk Perception Toward Avian Influenza Virus Exposure Among Cuban Hunters.Front Public Health. 2021 Jul 23;9:644786. doi: 10.3389/fpubh.2021.644786. eCollection 2021. Front Public Health. 2021. PMID: 34368040 Free PMC article.
References
LinkOut - more resources
Full Text Sources

