Rapid EGFR Mutation Detection Using the Idylla Platform: Single-Institution Experience of 1200 Cases Analyzed by an In-House Developed Pipeline and Comparison with Concurrent Next-Generation Sequencing Results
- PMID: 33346146
- PMCID: PMC7919857
- DOI: 10.1016/j.jmoldx.2020.11.009
Rapid EGFR Mutation Detection Using the Idylla Platform: Single-Institution Experience of 1200 Cases Analyzed by an In-House Developed Pipeline and Comparison with Concurrent Next-Generation Sequencing Results
Abstract
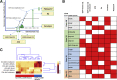
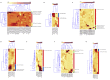
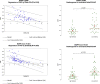
Mutations in the epidermal growth factor receptor (EGFR) are the most common targetable alterations in lung adenocarcinoma. To facilitate rapid testing, the Idylla EGFR assay was incorporated as a screening method before next-generation sequencing (NGS). Validation and experience using an in-house developed analysis pipeline, enhanced with a manual review algorithm is described. Results are compared with corresponding NGS results. In all, 1249 samples were studied. Validation demonstrated 98.57% (69/70) concordance with the reference methods. The limit of detection varied from 2% to 5% variant allele frequency for total EGFR quantitation cycle between 20 and 23. Of the 1179 clinical cases, 23.41% were EGFR-positive by Idylla. Concurrent NGS was successfully performed on 94.9% (799/842) requests. Concordance of Idylla with NGS was 98.62% (788/799) and 98.50% (787/799) using our in-house and Idylla analysis pipelines, respectively. Discordances involved missed mutations by both assays associated with low tumor/low input. Incorporating a manual review algorithm to supplement automated calls improved accuracy from 98.62% to 99.37% and sensitivity from 94.68% to 97.58%. Overall reporting time, from receipt of material to official clinical report, ranged from 1 to 3 days. Therefore, Idylla EGFR testing enables rapid and sensitive screening without compromising subsequent comprehensive NGS, when required. Automated calling, enhanced with a manual review algorithm, reduces false-negative calls associated with low tumor/low input samples.
Copyright © 2021 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Jordan E.J., Kim H.R., Arcila M.E., Barron D., Chakravarty D., Gao J., Chang M.T., Ni A., Kundra R., Jonsson P. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. - PMC - PubMed
-
- Lohinai Z., Hoda M.A., Fabian K., Ostoros G., Raso E., Barbai T., Timar J., Kovalszky I., Cserepes M., Rozsas A., Laszlo V., Grusch M., Berger W., Klepetko W., Moldvay J., Dome B., Hegedus B. Distinct epidemiology and clinical consequence of classic versus rare EGFR mutations in lung adenocarcinoma. J Thorac Oncol. 2015;10:738–746. - PubMed
-
- Dogan S., Shen R., Ang D.C., Johnson M.L., D'Angelo S.P., Paik P.K., Brzostowski E.B., Riely G.J., Kris M.G., Zakowski M.F., Ladanyi M. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18:6169–6177. - PMC - PubMed
-
- Shi Y., Au J.S., Thongprasert S., Srinivasan S., Tsai C.M., Khoa M.T., Heeroma K., Itoh Y., Cornelio G., Yang P.C. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9:154–162. - PMC - PubMed
-
- Lindeman N.I., Cagle P.T., Aisner D.L., Arcila M.E., Beasley M.B., Bernicker E.H., Colasacco C., Dacic S., Hirsch F.R., Kerr K., Kwiatkowski D.J., Ladanyi M., Nowak J.A., Sholl L., Temple-Smolkin R., Solomon B., Souter L.H., Thunnissen E., Tsao M.S., Ventura C.B., Wynes M.W., Yatabe Y. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn. 2018;20:129–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

