Evaluation of Homology-Independent CRISPR-Cas9 Off-Target Assessment Methods
- PMID: 33346710
- PMCID: PMC7757695
- DOI: 10.1089/crispr.2020.0053
Evaluation of Homology-Independent CRISPR-Cas9 Off-Target Assessment Methods
Erratum in
-
Correction to: Evaluation of Homology-Independent CRISPR-Cas9 Off-Target Assessment Methods, by Chaudhari et al. CRISPR J 2020; 3(6):440-453; DOI: 10.1089/chrispr.2020.0053.CRISPR J. 2021 Feb;4(1):155. doi: 10.1089/crispr.2020.0053.correx. CRISPR J. 2021. PMID: 33616443 Free PMC article. No abstract available.
Abstract
The ability to alter genomes specifically by CRISPR-Cas gene editing has revolutionized biological research, biotechnology, and medicine. Broad therapeutic application of this technology, however, will require thorough preclinical assessment of off-target editing by homology-based prediction coupled with reliable methods for detecting off-target editing. Several off-target site nomination assays exist, but careful comparison is needed to ascertain their relative strengths and weaknesses. In this study, HEK293T cells were treated with Streptococcus pyogenes Cas9 and eight guide RNAs with varying levels of predicted promiscuity in order to compare the performance of three homology-independent off-target nomination methods: the cell-based assay, GUIDE-seq, and the biochemical assays CIRCLE-seq and SITE-seq. The three methods were benchmarked by sequencing 75,000 homology-nominated sites using hybrid capture followed by high-throughput sequencing, providing the most comprehensive assessment of such methods to date. The three methods performed similarly in nominating sequence-confirmed off-target sites, but with large differences in the total number of sites nominated. When combined with homology-dependent nomination methods and confirmation by sequencing, all three off-target nomination methods provide a comprehensive assessment of off-target activity. GUIDE-seq's low false-positive rate and the high correlation of its signal with observed editing highlight its suitability for nominating off-target sites for ex vivo CRISPR-Cas therapies.
Figures

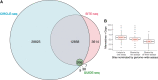

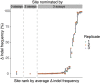

Comment in
-
Assessing Off-Target Editing of CRISPR-Cas9 Systems.CRISPR J. 2020 Dec;3(6):430-432. doi: 10.1089/crispr.2020.29116.smi. CRISPR J. 2020. PMID: 33346715 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
