Non-duplex G-Quadruplex DNA Structure: A Developing Story from Predicted Sequences to DNA Structure-Dependent Epigenetics and Beyond
- PMID: 33347280
- PMCID: PMC7612685
- DOI: 10.1021/acs.accounts.0c00431
Non-duplex G-Quadruplex DNA Structure: A Developing Story from Predicted Sequences to DNA Structure-Dependent Epigenetics and Beyond
Abstract
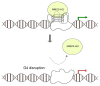
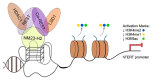
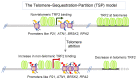
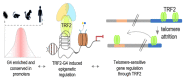
The story of the non-duplex DNA form known as the G-quadruplex (G4) has traversed a winding path. From initial skepticism followed by debate to a surge in interest, the G4 story intertwines many threads. Starting with computational predictions of a gene regulatory role, which now include epigenetic functions, our group was involved in many of these advances along with many other laboratories. Following a brief background, set in the latter half of the last century when the concept of the G4 as a structure took ground, here we account the developments. This is through a lens that though focused on our groups' research presents work from many other groups that played significant roles. Together these provide a broad perspective to the G4 story. Initially we were intrigued on seeing potential G4 (pG4)-forming sequences, then known to be found primarily at the telomeres and immunoglobin switch regions, occurring throughout the genome and being particularly prevalent in promoters of bacteria. We further observed that pG4s were not only prevalent but also conserved through evolution in promoters of human, chimpanzee, mouse and rat genomes. This was between 2005 and 2007. Encouraged by these partly and partly in response to the view held by many that genome-wide presence of G4s were genomic "accidents", the focus shifted to seeking experimental evidence.In the next year, 2008, two independent findings showed promise. First, on treating human cancer cells with G4-binding ligands, we observed widespread change in gene expression. Second, our search for the missing G4-specific transcription factor, without which, importantly, G4s in promoters posed only half the story, yielded results. We determined how NM23-H2 (also known as NME2 or NDPK-B) interacts with G4s and how interaction of NM23-H2 with a G4 in the promoter of the oncogene c-myc was important for regulation of c-myc transcription. NM23-H2, and subsequently many other similar factors discovered by multiple groups, is possibly giving shape to what might be the "G4-transcriptome". Later, a close look at NM23-H2-G4 interaction in regulation of the human reverse transcriptase gene (hTERT) revealed the role of G4s in local epigenetic modifications. Meanwhile work from others showed how G4s impact histone modifications following replication. Together these show the intrinsic role of DNA sequence, through formation of DNA structure, in epigenetics.More recent work, however, was waiting to reveal aspects that tend to bring forth a completely new understanding of G4s. We observed that the telomere-repeat-binding-factor-2 (TRF2), known canonically to be telomere-associated, binds extensively outside telomeres throughout the genome. Moreover, a large fraction of the non-telomeric TRF2 sites comprise G4s. Second, the extent of non-telomeric TRF2 binding at promoters was dependent on telomere length. Thereby TRF2-induced epigenetic gene regulation was telomere-dependent. Together these implicate underlying connections that show signs of addressing an intriguing unanswered question that takes us back to the beginning: Why are G4s prevalent in two distinct regions, the telomeres and gene promoters?
Conflict of interest statement
Notes
The authors declare no competing financial interest.
Figures



References
-
- Mukherjee AK, Sharma S, Bagri S, Kutum R, Kumar P, Hussain A, Singh P, Saha D, Kar A, Dash D, Chowdhury S. Telomere Repeat-Binding Factor 2 Binds Extensively to Extra-Telomeric G-Quadruplexes and Regulates the Epigenetic Status of Several Gene Promoters. J Biol Chem. 2019;294:17709–17722. - PMC - PubMed
-
- Zimmerman SB, Cohen GH, Davies DR. X-Ray Fiber Diffraction and Model-Building Study of Polyguanylic Acid and Polyinosinic Acid. J Mol Biol. 1975;92:181–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

