Toward a scalable framework for reproducible processing of volumetric, nanoscale neuroimaging datasets
- PMID: 33347572
- PMCID: PMC7751400
- DOI: 10.1093/gigascience/giaa147
Toward a scalable framework for reproducible processing of volumetric, nanoscale neuroimaging datasets
Abstract
Background: Emerging neuroimaging datasets (collected with imaging techniques such as electron microscopy, optical microscopy, or X-ray microtomography) describe the location and properties of neurons and their connections at unprecedented scale, promising new ways of understanding the brain. These modern imaging techniques used to interrogate the brain can quickly accumulate gigabytes to petabytes of structural brain imaging data. Unfortunately, many neuroscience laboratories lack the computational resources to work with datasets of this size: computer vision tools are often not portable or scalable, and there is considerable difficulty in reproducing results or extending methods.
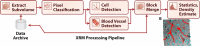
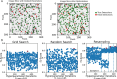
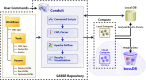
Results: We developed an ecosystem of neuroimaging data analysis pipelines that use open-source algorithms to create standardized modules and end-to-end optimized approaches. As exemplars we apply our tools to estimate synapse-level connectomes from electron microscopy data and cell distributions from X-ray microtomography data. To facilitate scientific discovery, we propose a generalized processing framework, which connects and extends existing open-source projects to provide large-scale data storage, reproducible algorithms, and workflow execution engines.
Conclusions: Our accessible methods and pipelines demonstrate that approaches across multiple neuroimaging experiments can be standardized and applied to diverse datasets. The techniques developed are demonstrated on neuroimaging datasets but may be applied to similar problems in other domains.
Keywords: computational neuroscience; containers; electron microscopy; microtomography; optimization; reproducible science; workflows.
© The Author(s) 2020. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Micheva KD, O’Rourke N, Busse B, et al. Array tomography: High-resolution three-dimensional immunofluorescence. Cold Spring Harb Protoc. 2010;5(11):1214–9. - PubMed
-
- Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat Methods. 2013;10(6):508–13. - PubMed
-
- Allen Institute for Brain Science. Allen Brain Atlas. http://brain-map.org/api/index.html. Accessed on June 6, 2018.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

