Lung Adenocarcinoma Mouse Models Based on Orthotopic Transplantation of Syngeneic Tumor-Initiating Cells Expressing EpCAM, SCA-1, and Ly6d
- PMID: 33348616
- PMCID: PMC7767274
- DOI: 10.3390/cancers12123805
Lung Adenocarcinoma Mouse Models Based on Orthotopic Transplantation of Syngeneic Tumor-Initiating Cells Expressing EpCAM, SCA-1, and Ly6d
Abstract
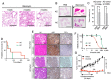
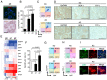
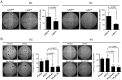
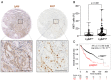
Somatic mutations in EGFR and KRAS as well as chromosome rearrangements affecting ALK, ROS1, and RET have been identified in human lung adenocarcinoma (LUAD). We here developed organoid-based orthotopic and syngeneic mouse models for studies of the pathogenesis and treatment of LUAD. We isolated EpCAM-positive epithelial cells from mouse lungs and cultured them as organoids to maintain epithelial stem cell properties. These cells were transformed by KRAS(G12V) or EML4-ALK and then transplanted via the trachea into the lungs of the syngeneic mice, where they formed tumors that expressed the lung lineage marker TTF-1 and which closely recapitulated the pathology of human LUAD. Treatment with crizotinib suppressed the growth of tumors formed by the EML4-ALK-expressing lung epithelial cells in a subcutaneous transplantation model. Organoid culture of normal lung epithelial cells resulted in enrichment of EpCAM+SCA-1(Ly6a)+ cells as well as in that of cells expressing another member of the Ly6 protein family, Ly6d, which was found to be required for the growth of the LUAD-initiating cells expressing KRAS(G12V) or EML4-ALK. We also found that a high expression level of LY6D was associated with poor prognosis in human LUAD. Our results thus suggest that LY6D is a potential lung cancer stem cell marker.
Keywords: EpCAM; Ly6d; SCA-1; lung cancer; orthotopic transplantation; syngeneic mouse model.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

