The Structure and Function of Paraoxonase-1 and Its Comparison to Paraoxonase-2 and -3
- PMID: 33348669
- PMCID: PMC7766523
- DOI: 10.3390/molecules25245980
The Structure and Function of Paraoxonase-1 and Its Comparison to Paraoxonase-2 and -3
Abstract
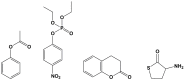
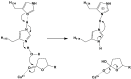
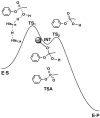
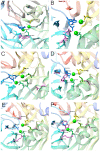
Serum paraoxonase-1 (PON1) is the most studied member of the group of paraoxonases (PONs). This enzyme possesses three enzymatic activities: lactonase, arylesterase, and paraoxonase activity. PON1 and its isoforms play an important role in drug metabolism as well as in the prevention of cardiovascular and neurodegenerative diseases. Although all three members of the PON family have the same origin and very similar amino acid sequences, they have different functions and are found in different locations. PONs exhibit substrate promiscuity, and their true physiological substrates are still not known. However, possible substrates include homocysteine thiolactone, an analogue of natural quorum-sensing molecules, and the recently discovered derivatives of arachidonic acid-bioactive δ-lactones. Directed evolution, site-directed mutagenesis, and kinetic studies provide comprehensive insights into the active site and catalytic mechanism of PON1. However, there is still a whole world of mystery waiting to be discovered, which would elucidate the substrate promiscuity of a group of enzymes that are so similar in their evolution and sequence yet so distinct in their function.
Keywords: PON1; PON2; PON3; arylesterase; atherosclerosis; kinetic; lactonase; organophosphate; oxidative stress; paraoxonase; structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Paraoxonases-1, -2 and -3: What are their functions?Chem Biol Interact. 2016 Nov 25;259(Pt B):51-62. doi: 10.1016/j.cbi.2016.05.036. Epub 2016 May 26. Chem Biol Interact. 2016. PMID: 27238723 Free PMC article.
-
Identification of biologically active δ-lactone eicosanoids as paraoxonase substrates.Biochem Biophys Res Commun. 2018 Oct 20;505(1):87-92. doi: 10.1016/j.bbrc.2018.09.083. Epub 2018 Sep 18. Biochem Biophys Res Commun. 2018. PMID: 30241945
-
Characterization of human paraoxonase 1 variants suggest that His residues at 115 and 134 positions are not always needed for the lactonase/arylesterase activities of the enzyme.Protein Sci. 2013 Dec;22(12):1799-807. doi: 10.1002/pro.2380. Epub 2013 Oct 26. Protein Sci. 2013. PMID: 24123308 Free PMC article.
-
The human paraoxonase gene cluster as a target in the treatment of atherosclerosis.Antioxid Redox Signal. 2012 Mar 15;16(6):597-632. doi: 10.1089/ars.2010.3774. Epub 2011 Oct 18. Antioxid Redox Signal. 2012. PMID: 21867409 Free PMC article. Review.
-
The paraoxonase 1, 2 and 3 in humans.Biochem Med (Zagreb). 2011;21(2):122-30. doi: 10.11613/bm.2011.020. Biochem Med (Zagreb). 2011. PMID: 22135851 Review.
Cited by
-
Pulsed Electromagnetic Fields Induce Skeletal Muscle Cell Repair by Sustaining the Expression of Proteins Involved in the Response to Cellular Damage and Oxidative Stress.Int J Mol Sci. 2023 Nov 23;24(23):16631. doi: 10.3390/ijms242316631. Int J Mol Sci. 2023. PMID: 38068954 Free PMC article.
-
Modulatory Effect of Lifestyle-Related, Environmental and Genetic Factors on Paraoxonase-1 Activity: A Review.Int J Environ Res Public Health. 2023 Feb 5;20(4):2813. doi: 10.3390/ijerph20042813. Int J Environ Res Public Health. 2023. PMID: 36833509 Free PMC article. Review.
-
Association between human paraoxonase 2 protein and efficacy of acetylcholinesterase inhibiting drugs used against Alzheimer's disease.PLoS One. 2021 Oct 29;16(10):e0258879. doi: 10.1371/journal.pone.0258879. eCollection 2021. PLoS One. 2021. PMID: 34714861 Free PMC article.
-
Vutiglabridin Modulates Paraoxonase 1 and Ameliorates Diet-Induced Obesity in Hyperlipidemic Mice.Biomolecules. 2023 Apr 18;13(4):687. doi: 10.3390/biom13040687. Biomolecules. 2023. PMID: 37189434 Free PMC article.
-
Association between Paraoxonase/Arylesterase Activity of Serum PON-1 Enzyme and Rheumatoid Arthritis: A Systematic Review and Meta-Analysis.Antioxidants (Basel). 2022 Nov 23;11(12):2317. doi: 10.3390/antiox11122317. Antioxidants (Basel). 2022. PMID: 36552525 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

