Physiological, Biochemical, and Transcriptomic Responses of Neolamarckia cadamba to Aluminum Stress
- PMID: 33348765
- PMCID: PMC7767006
- DOI: 10.3390/ijms21249624
Physiological, Biochemical, and Transcriptomic Responses of Neolamarckia cadamba to Aluminum Stress
Abstract
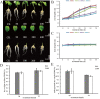
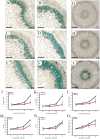
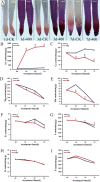
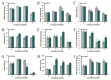
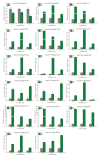
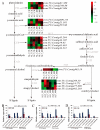
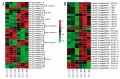
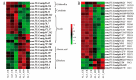
Aluminum is the most abundant metal of the Earth's crust accounting for 7% of its mass, and release of toxic Al3+ in acid soils restricts plant growth. Neolamarckia cadamba, a fast-growing tree, only grows in tropical regions with acidic soils. In this study, N. cadamba was treated with high concentrations of aluminum under acidic condition (pH 4.5) to study its physiological, biochemical, and molecular response mechanisms against high aluminum stress. High aluminum concentration resulted in significant inhibition of root growth with time in N. cadamba. The concentration of Al3+ ions in the root tip increased significantly and the distribution of absorbed Al3+ was observed in the root tip after Al stress. Meanwhile, the concentration of Ca, Mg, Mn, and Fe was significantly decreased, but P concentration increased. Aluminum stress increased activities of antioxidant enzymes such as superoxide dismutase (SOD), catalase from micrococcus lysodeiktic (CAT), and peroxidase (POD) in the root tip, while the content of MDA was decreased. Transcriptome analysis showed 37,478 differential expression genes (DEGs) and 4096 GOs terms significantly associated with treatments. The expression of genes regulating aluminum transport and abscisic acid synthesis was significantly upregulated; however, the genes involved in auxin synthesis were downregulated. Of note, the transcripts of several key enzymes affecting lignin monomer synthesis in phenylalanine pathway were upregulated. Our results shed light on the physiological and molecular mechanisms of aluminum stress tolerance in N. cadamba.
Keywords: DEGs; Neolamarckia cadamba; ROS; aluminum stress; hormone; signal transduction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Hoekenga O.A., Vision T.J., Shaff J.E., Monforte A.J., Lee G.P., Howell S.H., Kochian L.V. Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta x Columbia) by quantitative trait locus mapping. A physiologically simple but genetically complex trait. Plant Physiol. 2003;132:936–948. doi: 10.1104/pp.103.023085. - DOI - PMC - PubMed
-
- Samac D.A., Tesfaye M. Plant improvement for tolerance to aluminum in acid soils—A review. Plant Cell Tissue Organ Cult. (PCTOC) 2003;75:189–207. doi: 10.1023/A:1025843829545. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

