Structural Determinants and Their Role in Cyanobacterial Morphogenesis
- PMID: 33348886
- PMCID: PMC7766704
- DOI: 10.3390/life10120355
Structural Determinants and Their Role in Cyanobacterial Morphogenesis
Abstract
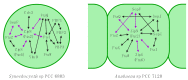
Cells have to erect and sustain an organized and dynamically adaptable structure for an efficient mode of operation that allows drastic morphological changes during cell growth and cell division. These manifold tasks are complied by the so-called cytoskeleton and its associated proteins. In bacteria, FtsZ and MreB, the bacterial homologs to tubulin and actin, respectively, as well as coiled-coil-rich proteins of intermediate filament (IF)-like function to fulfil these tasks. Despite generally being characterized as Gram-negative, cyanobacteria have a remarkably thick peptidoglycan layer and possess Gram-positive-specific cell division proteins such as SepF and DivIVA-like proteins, besides Gram-negative and cyanobacterial-specific cell division proteins like MinE, SepI, ZipN (Ftn2) and ZipS (Ftn6). The diversity of cellular morphologies and cell growth strategies in cyanobacteria could therefore be the result of additional unidentified structural determinants such as cytoskeletal proteins. In this article, we review the current advances in the understanding of the cyanobacterial cell shape, cell division and cell growth.
Keywords: FtsZ; IF proteins; MreB; cell division; cell shape; cyanobacteria; cytoskeleton; morphology.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Alberts B., Johnson A., Lewis J., Morgan D., Raff M., Roberts K., Walter P. Molecular Biology of the Cell. 6th ed. Garland Science; New York, NY, USA: 2014.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

