The diversity of Shine-Dalgarno sequences sheds light on the evolution of translation initiation
- PMID: 33349119
- PMCID: PMC8583243
- DOI: 10.1080/15476286.2020.1861406
The diversity of Shine-Dalgarno sequences sheds light on the evolution of translation initiation
Abstract
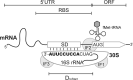
Shine-Dalgarno (SD) sequences, the core element of prokaryotic ribosome-binding sites, facilitate mRNA translation by base-pair interaction with the anti-SD (aSD) sequence of 16S rRNA. In contrast to this paradigm, an inspection of thousands of prokaryotic species unravels tremendous SD sequence diversity both within and between genomes, whereas aSD sequences remain largely static. The pattern has led many to suggest unidentified mechanisms for translation initiation. Here we review known translation-initiation pathways in prokaryotes. Moreover, we seek to understand the cause and consequence of SD diversity through surveying recent advances in biochemistry, genomics, and high-throughput genetics. These findings collectively show: (1) SD:aSD base pairing is beneficial but nonessential to translation initiation. (2) The 5' untranslated region of mRNA evolves dynamically and correlates with organismal phylogeny and ecological niches. (3) Ribosomes have evolved distinct usage of translation-initiation pathways in different species. We propose a model portraying the SD diversity shaped by optimization of gene expression, adaptation to environments and growth demands, and the species-specific prerequisite of ribosomes to initiate translation. The model highlights the coevolution of ribosomes and mRNA features, leading to functional customization of the translation apparatus in each organism.
Keywords: Shine-Dalgarno sequence; diversity; evolution; mRNA; ribosome; transcriptome; translation initiation.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures



References
-
- Milon P, Konevega AL, Gualerzi CO, et al. Kinetic checkpoint at a late step in translation initiation. Mol Cell. 2008;30:712–720. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
