A variant-centric perspective on geographic patterns of human allele frequency variation
- PMID: 33350384
- PMCID: PMC7755386
- DOI: 10.7554/eLife.60107
A variant-centric perspective on geographic patterns of human allele frequency variation
Abstract
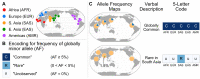
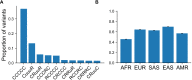
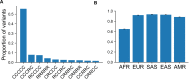
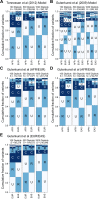
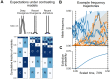
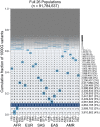
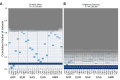
A key challenge in human genetics is to understand the geographic distribution of human genetic variation. Often genetic variation is described by showing relationships among populations or individuals, drawing inferences over many variants. Here, we introduce an alternative representation of genetic variation that reveals the relative abundance of different allele frequency patterns. This approach allows viewers to easily see several features of human genetic structure: (1) most variants are rare and geographically localized, (2) variants that are common in a single geographic region are more likely to be shared across the globe than to be private to that region, and (3) where two individuals differ, it is most often due to variants that are found globally, regardless of whether the individuals are from the same region or different regions. Our variant-centric visualization clarifies the geographic patterns of human variation and can help address misconceptions about genetic differentiation among populations.
Keywords: data visualization; demographic history; evolutionary biology; genetics; genomics; human; human variation; population genetics; population structure.
© 2020, Biddanda et al.
Conflict of interest statement
AB, DR, JN No competing interests declared
Figures










Similar articles
-
Admixture and the organization of genetic diversity in a butterfly species complex revealed through common and rare genetic variants.Mol Ecol. 2014 Sep;23(18):4555-73. doi: 10.1111/mec.12811. Epub 2014 Jun 16. Mol Ecol. 2014. PMID: 24866941
-
Human Disease Variation in the Light of Population Genomics.Cell. 2019 Mar 21;177(1):115-131. doi: 10.1016/j.cell.2019.01.052. Cell. 2019. PMID: 30901534 Review.
-
The global spectrum of protein-coding pharmacogenomic diversity.Pharmacogenomics J. 2018 Jan;18(1):187-195. doi: 10.1038/tpj.2016.77. Epub 2016 Oct 25. Pharmacogenomics J. 2018. PMID: 27779249 Free PMC article.
-
Genetics in geographically structured populations: defining, estimating and interpreting F(ST).Nat Rev Genet. 2009 Sep;10(9):639-50. doi: 10.1038/nrg2611. Nat Rev Genet. 2009. PMID: 19687804 Free PMC article. Review.
-
Population genetic structuring in Acanthopagrus butcheri (Pisces: Sparidae): does low gene flow among estuaries apply to both sexes?Mar Biotechnol (NY). 2007 Jan-Feb;9(1):33-44. doi: 10.1007/s10126-006-6023-7. Epub 2006 Aug 22. Mar Biotechnol (NY). 2007. PMID: 16937020
Cited by
-
Beyond the Human Genome Project: The Age of Complete Human Genome Sequences and Pangenome References.Annu Rev Genomics Hum Genet. 2024 Aug;25(1):77-104. doi: 10.1146/annurev-genom-021623-081639. Epub 2024 Aug 6. Annu Rev Genomics Hum Genet. 2024. PMID: 38663087 Free PMC article. Review.
-
The background and legacy of Lewontin's apportionment of human genetic diversity.Philos Trans R Soc Lond B Biol Sci. 2022 Jun 6;377(1852):20200406. doi: 10.1098/rstb.2020.0406. Epub 2022 Apr 18. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35430890 Free PMC article. Review.
-
The distribution of highly deleterious variants across human ancestry groups.Proc Natl Acad Sci U S A. 2025 May 27;122(21):e2503857122. doi: 10.1073/pnas.2503857122. Epub 2025 May 23. Proc Natl Acad Sci U S A. 2025. PMID: 40408403
-
Similarity-Based Analysis of Allele Frequency Distribution among Multiple Populations Identifies Adaptive Genomic Structural Variants.Mol Biol Evol. 2022 Mar 2;39(3):msab313. doi: 10.1093/molbev/msab313. Mol Biol Evol. 2022. PMID: 34718708 Free PMC article.
-
Imputation accuracy across global human populations.Am J Hum Genet. 2024 May 2;111(5):979-989. doi: 10.1016/j.ajhg.2024.03.011. Epub 2024 Apr 10. Am J Hum Genet. 2024. PMID: 38604166 Free PMC article.
References
-
- Adrion JR, Cole CB, Dukler N, Galloway JG, Gladstein AL, Gower G, Kyriazis CC, Ragsdale AP, Tsambos G, Baumdicker F, Carlson J, Cartwright RA, Durvasula A, Gronau I, Kim BY, McKenzie P, Messer PW, Noskova E, Ortega-Del Vecchyo D, Racimo F, Struck TJ, Gravel S, Gutenkunst RN, Lohmueller KE, Ralph PL, Schrider DR, Siepel A, Kelleher J, Kern AD. A community-maintained standard library of population genetic models. eLife. 2020;9:e54967. doi: 10.7554/eLife.54967. - DOI - PMC - PubMed
-
- Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, Chen Y, Felkel S, Hallast P, Kamm J, Blanché H, Deleuze J-F, Cann H, Mallick S, Reich D, Sandhu MS, Skoglund P, Scally A, Xue Y, Durbin R, Tyler-Smith C. Insights into human genetic variation and population history from 929 diverse genomes. bioRxiv. 2019 doi: 10.1101/674986. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

