Individual differences in honey bee behavior enabled by plasticity in brain gene regulatory networks
- PMID: 33350385
- PMCID: PMC7755388
- DOI: 10.7554/eLife.62850
Individual differences in honey bee behavior enabled by plasticity in brain gene regulatory networks
Abstract
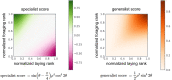
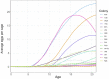
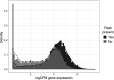
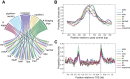
Understanding the regulatory architecture of phenotypic variation is a fundamental goal in biology, but connections between gene regulatory network (GRN) activity and individual differences in behavior are poorly understood. We characterized the molecular basis of behavioral plasticity in queenless honey bee (Apis mellifera) colonies, where individuals engage in both reproductive and non-reproductive behaviors. Using high-throughput behavioral tracking, we discovered these colonies contain a continuum of phenotypes, with some individuals specialized for either egg-laying or foraging and 'generalists' that perform both. Brain gene expression and chromatin accessibility profiles were correlated with behavioral variation, with generalists intermediate in behavior and molecular profiles. Models of brain GRNs constructed for individuals revealed that transcription factor (TF) activity was highly predictive of behavior, and behavior-associated regulatory regions had more TF motifs. These results provide new insights into the important role played by brain GRN plasticity in the regulation of behavior, with implications for social evolution.
Keywords: Apis mellifera; behavioral plasticity; ecology; gene regulation; genetics; genomics.
© 2020, Jones et al.
Conflict of interest statement
BJ, VR, TG, TJ, AC, BR, TC, SB, SH, CB, MM, SS, SC, GR No competing interests declared
Figures






References
-
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Wicke M, Yu Y, Zheng X, Brain G, Osdi I, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Wicke M, Yu Y, Zheng X. TensorFlow : a system for large-scale machine learning. OSDI. 2016;16:265–283. doi: 10.1126/science.aab4113.4. - DOI
-
- Ament SA, Wang Y, Chen CC, Blatti CA, Hong F, Liang ZS, Negre N, White KP, Rodriguez-Zas SL, Mizzen CA, Sinha S, Zhong S, Robinson GE. The transcription factor ultraspiracle influences honey bee social behavior and behavior-related gene expression. PLOS Genetics. 2012;8:e1002596. doi: 10.1371/journal.pgen.1002596. - DOI - PMC - PubMed
-
- Araya CL, Kawli T, Kundaje A, Jiang L, Wu B, Vafeados D, Terrell R, Weissdepp P, Gevirtzman L, Mace D, Niu W, Boyle AP, Xie D, Ma L, Murray JI, Reinke V, Waterston RH, Snyder M. Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature. 2014;512:400–405. doi: 10.1038/nature13497. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

