The role of disorder in RNA binding affinity and specificity
- PMID: 33352065
- PMCID: PMC7776568
- DOI: 10.1098/rsob.200328
The role of disorder in RNA binding affinity and specificity
Abstract
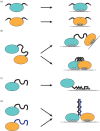
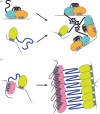
Most RNA-binding modules are small and bind few nucleotides. RNA-binding proteins typically attain the physiological specificity and affinity for their RNA targets by combining several RNA-binding modules. Here, we review how disordered linkers connecting RNA-binding modules govern the specificity and affinity of RNA-protein interactions by regulating the effective concentration of these modules and their relative orientation. RNA-binding proteins also often contain extended intrinsically disordered regions that mediate protein-protein and RNA-protein interactions with multiple partners. We discuss how these regions can connect proteins and RNA resulting in heterogeneous higher-order assemblies such as membrane-less compartments and amyloid-like structures that have the characteristics of multi-modular entities. The assembled state generates additional RNA-binding specificity and affinity properties that contribute to further the function of RNA-binding proteins within the cellular environment.
Keywords: RNA-binding domains; RNA-binding modules; RNA-binding proteins; amyloids; assemblies; intrinsically disordered regions; linkers.
Conflict of interest statement
The authors declare no competing interests.
Figures
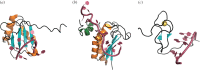
References
-
- Lewin B, Krebs JE, Goldstein ES, Kilpatrick ST. 2018. Lewin's genes XII, 12th edn Burlington, MA: Jones and Bartlett Learning.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

