Population studies of the wild tomato species Solanum chilense reveal geographically structured major gene-mediated pathogen resistance
- PMID: 33352079
- PMCID: PMC7779489
- DOI: 10.1098/rspb.2020.2723
Population studies of the wild tomato species Solanum chilense reveal geographically structured major gene-mediated pathogen resistance
Abstract
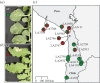
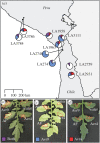
Natural plant populations encounter strong pathogen pressure and defence-associated genes are known to be under selection dependent on the pressure by the pathogens. Here, we use populations of the wild tomato Solanum chilense to investigate natural resistance against Cladosporium fulvum, a well-known ascomycete pathogen of domesticated tomatoes. Host populations used are from distinct geographical origins and share a defined evolutionary history. We show that distinct populations of S. chilense differ in resistance against the pathogen. Screening for major resistance gene-mediated pathogen recognition throughout the whole species showed clear geographical differences between populations and complete loss of pathogen recognition in the south of the species range. In addition, we observed high complexity in a homologues of Cladosporium resistance (Hcr) locus, underlying the recognition of C. fulvum, in central and northern populations. Our findings show that major gene-mediated recognition specificity is diverse in a natural plant-pathosystem. We place major gene resistance in a geographical context that also defined the evolutionary history of that species. Data suggest that the underlying loci are more complex than previously anticipated, with small-scale gene recombination being possibly responsible for maintaining balanced polymorphisms in the populations that experience pathogen pressure.
Keywords: Cladosporium fulvum; Solanum chilense; loss of resistance; major gene resistance; tomato.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures


Similar articles
-
Recognitional specificity and evolution in the tomato-Cladosporium fulvum pathosystem.Mol Plant Microbe Interact. 2009 Oct;22(10):1191-202. doi: 10.1094/MPMI-22-10-1191. Mol Plant Microbe Interact. 2009. PMID: 19737093 Review.
-
Physiological and RNA-seq analyses provide insights into the response mechanism of the Cf-10-mediated resistance to Cladosporium fulvum infection in tomato.Plant Mol Biol. 2018 Mar;96(4-5):403-416. doi: 10.1007/s11103-018-0706-0. Epub 2018 Jan 30. Plant Mol Biol. 2018. PMID: 29383477
-
Genes Encoding Recognition of the Cladosporium fulvum Effector Protein Ecp5 Are Encoded at Several Loci in the Tomato Genome.G3 (Bethesda). 2020 May 4;10(5):1753-1763. doi: 10.1534/g3.120.401119. G3 (Bethesda). 2020. PMID: 32209596 Free PMC article.
-
Analysis of five near-complete genome assemblies of the tomato pathogen Cladosporium fulvum uncovers additional accessory chromosomes and structural variations induced by transposable elements effecting the loss of avirulence genes.BMC Biol. 2024 Jan 29;22(1):25. doi: 10.1186/s12915-024-01818-z. BMC Biol. 2024. PMID: 38281938 Free PMC article.
-
Molecular communication between host plant and the fungal tomato pathogen Cladosporium fulvum.Antonie Van Leeuwenhoek. 1994;65(3):257-62. doi: 10.1007/BF00871954. Antonie Van Leeuwenhoek. 1994. PMID: 7847893 Review.
Cited by
-
Unveiling diversity and adaptations of the wild tomato Microbiome in their center of origin in the Ecuadorian Andes.Sci Rep. 2025 Jul 1;15(1):22448. doi: 10.1038/s41598-025-05816-1. Sci Rep. 2025. PMID: 40594354 Free PMC article.
-
Multi-omics approach highlights differences between RLP classes in Arabidopsis thaliana.BMC Genomics. 2021 Jul 20;22(1):557. doi: 10.1186/s12864-021-07855-0. BMC Genomics. 2021. PMID: 34284718 Free PMC article.
-
Beyond the genomes of Fulvia fulva (syn. Cladosporium fulvum) and Dothistroma septosporum: New insights into how these fungal pathogens interact with their host plants.Mol Plant Pathol. 2023 May;24(5):474-494. doi: 10.1111/mpp.13309. Epub 2023 Feb 15. Mol Plant Pathol. 2023. PMID: 36790136 Free PMC article. Review.
-
Patterns of presence-absence variation of NLRs across populations of Solanum chilense are clade-dependent and mainly shaped by past demographic history.New Phytol. 2025 Feb;245(4):1718-1732. doi: 10.1111/nph.20293. Epub 2024 Nov 24. New Phytol. 2025. PMID: 39582196 Free PMC article.
-
Laminarin-triggered defence responses are geographically dependent in natural populations of Solanum chilense.J Exp Bot. 2023 May 19;74(10):3240-3254. doi: 10.1093/jxb/erad087. J Exp Bot. 2023. PMID: 36880316 Free PMC article.
References
Publication types
MeSH terms
Supplementary concepts
Associated data
LinkOut - more resources
Full Text Sources

