Structural basis of client specificity in mitochondrial membrane-protein chaperones
- PMID: 33355130
- PMCID: PMC11206218
- DOI: 10.1126/sciadv.abd0263
Structural basis of client specificity in mitochondrial membrane-protein chaperones
Abstract
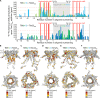
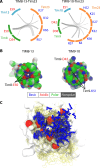
Chaperones are essential for assisting protein folding and for transferring poorly soluble proteins to their functional locations within cells. Hydrophobic interactions drive promiscuous chaperone-client binding, but our understanding of how additional interactions enable client specificity is sparse. Here, we decipher what determines binding of two chaperones (TIM8·13 and TIM9·10) to different integral membrane proteins, the all-transmembrane mitochondrial carrier Ggc1 and Tim23, which has an additional disordered hydrophilic domain. Combining NMR, SAXS, and molecular dynamics simulations, we determine the structures of Tim23/TIM8·13 and Tim23/TIM9·10 complexes. TIM8·13 uses transient salt bridges to interact with the hydrophilic part of its client, but its interactions to the transmembrane part are weaker than in TIM9·10. Consequently, TIM9·10 outcompetes TIM8·13 in binding hydrophobic clients, while TIM8·13 is tuned to few clients with both hydrophilic and hydrophobic parts. Our study exemplifies how chaperones fine-tune the balance of promiscuity versus specificity.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures




References
-
- Powers E. T., Morimoto R. I., Dillin A., Kelly J. W., Balch W. E., Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78, 959–991 (2009). - PubMed
-
- Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., Hartl F. U., Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 (2013). - PubMed
-
- Kim D. H., Lee J.-E., Xu Z.-Y., Geem K. R., Kwon Y., Park J. W., Hwang I., Cytosolic targeting factor AKR2A captures chloroplast outer membrane-localized client proteins at the ribosome during translation. Nat. Commun. 6, 6843 (2015). - PubMed
-
- Bose D., Chakrabarti A., Substrate specificity in the context of molecular chaperones. IUBMB Life 69, 647–659 (2017). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

