CRL4DCAF1/VprBP E3 ubiquitin ligase controls ribosome biogenesis, cell proliferation, and development
- PMID: 33355139
- PMCID: PMC11206221
- DOI: 10.1126/sciadv.abd6078
CRL4DCAF1/VprBP E3 ubiquitin ligase controls ribosome biogenesis, cell proliferation, and development
Abstract
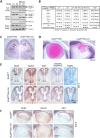
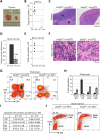
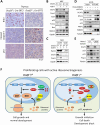
Evolutionarily conserved DCAF1 is a major substrate receptor for the DDB1-CUL4-ROC1 E3 ubiquitin ligase (CRL4) and controls cell proliferation and development. The molecular basis for these functions is unclear. We show here that DCAF1 loss in multiple tissues and organs selectively eliminates proliferating cells and causes perinatal lethality, thymic atrophy, and bone marrow defect. Inducible DCAF1 loss eliminates proliferating, but not quiescent, T cells and MEFs. We identify the ribosome assembly factor PWP1 as a substrate of the CRL4DCAF1 ligase. DCAF1 loss results in PWP1 accumulation, impairing rRNA processing and ribosome biogenesis. Knockdown or overexpression of PWP1 can rescue defects or cause similar defects as DCAF1 loss, respectively, in ribosome biogenesis. DCAF1 loss increases free RPL11, resulting in L11-MDM2 association and p53 activation. Cumulatively, these results reveal a critical function for DCAF1 in ribosome biogenesis and define a molecular basis of DCAF1 function in cell proliferation and development.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



Similar articles
-
The CRL4DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.EMBO J. 2021 Nov 15;40(22):e108008. doi: 10.15252/embj.2021108008. Epub 2021 Sep 30. EMBO J. 2021. PMID: 34595758 Free PMC article.
-
Inhibition of Vpx-Mediated SAMHD1 and Vpr-Mediated Host Helicase Transcription Factor Degradation by Selective Disruption of Viral CRL4 (DCAF1) E3 Ubiquitin Ligase Assembly.J Virol. 2017 Apr 13;91(9):e00225-17. doi: 10.1128/JVI.00225-17. Print 2017 May 1. J Virol. 2017. PMID: 28202763 Free PMC article.
-
Interactions with DCAF1 and DDB1 in the CRL4 E3 ubiquitin ligase are required for Vpr-mediated G2 arrest.Virol J. 2014 Jun 9;11:108. doi: 10.1186/1743-422X-11-108. Virol J. 2014. PMID: 24912982 Free PMC article.
-
VprBP (DCAF1): a promiscuous substrate recognition subunit that incorporates into both RING-family CRL4 and HECT-family EDD/UBR5 E3 ubiquitin ligases.BMC Mol Biol. 2013 Sep 13;14:22. doi: 10.1186/1471-2199-14-22. BMC Mol Biol. 2013. PMID: 24028781 Free PMC article. Review.
-
The role and mechanism of CRL4 E3 ubiquitin ligase in cancer and its potential therapy implications.Oncotarget. 2015 Dec 15;6(40):42590-602. doi: 10.18632/oncotarget.6052. Oncotarget. 2015. PMID: 26460955 Free PMC article. Review.
Cited by
-
Generation of nanoscopic membrane curvature for membrane trafficking.Nat Rev Mol Cell Biol. 2023 Jan;24(1):63-78. doi: 10.1038/s41580-022-00511-9. Epub 2022 Aug 2. Nat Rev Mol Cell Biol. 2023. PMID: 35918535 Review.
-
Ddb1 Is Essential for the Expansion of CD4+ Helper T Cells by Regulating Cell Cycle Progression and Cell Death.Front Immunol. 2021 Aug 30;12:722273. doi: 10.3389/fimmu.2021.722273. eCollection 2021. Front Immunol. 2021. PMID: 34526995 Free PMC article.
-
DCAF1 interacts with PARD3 to promote hepatocellular carcinoma progression and metastasis by activating the Akt signaling pathway.J Exp Clin Cancer Res. 2024 May 6;43(1):136. doi: 10.1186/s13046-024-03055-2. J Exp Clin Cancer Res. 2024. PMID: 38711082 Free PMC article.
-
Inhibition of EED-mediated histone methylation alleviates neuroinflammation by suppressing WNT-mediated dendritic cell migration.J Neuroinflammation. 2025 Apr 1;22(1):97. doi: 10.1186/s12974-025-03429-z. J Neuroinflammation. 2025. PMID: 40169990 Free PMC article.
-
PWP1 transcriptionally regulates p53, modulating apoptosis and cell cycle to promote gastric cancer progression.Apoptosis. 2025 Apr;30(3-4):693-709. doi: 10.1007/s10495-024-02049-x. Epub 2024 Dec 25. Apoptosis. 2025. PMID: 39720977 Free PMC article.
References
-
- Warner J. R., The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 (1999). - PubMed
-
- Lambertsson A., The Minute genes in Drosophila and their molecular functions. Adv. Genet. 38, 69–134 (1998). - PubMed
-
- Teng T., Thomas G., Mercer C. A., Growth control and ribosomopathies. Curr. Opin. Genet. Dev. 23, 63–71 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

