Autophagy Contributes to the Quality Control of Leaf Mitochondria
- PMID: 33355344
- PMCID: PMC8112837
- DOI: 10.1093/pcp/pcaa162
Autophagy Contributes to the Quality Control of Leaf Mitochondria
Abstract
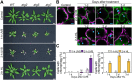
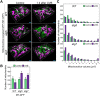
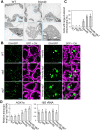
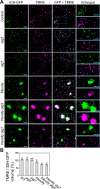
In autophagy, cytoplasmic components of eukaryotic cells are transported to lysosomes or the vacuole for degradation. Autophagy is involved in plant tolerance to the photooxidative stress caused by ultraviolet B (UVB) radiation, but its roles in plant adaptation to UVB damage have not been fully elucidated. Here, we characterized organellar behavior in UVB-damaged Arabidopsis (Arabidopsis thaliana) leaves and observed the occurrence of autophagic elimination of dysfunctional mitochondria, a process termed mitophagy. Notably, Arabidopsis plants blocked in autophagy displayed increased leaf chlorosis after a 1-h UVB exposure compared to wild-type plants. We visualized autophagosomes by labeling with a fluorescent protein-tagged autophagosome marker, AUTOPHAGY8 (ATG8), and found that a 1-h UVB treatment led to increased formation of autophagosomes and the active transport of mitochondria into the central vacuole. In atg mutant plants, the mitochondrial population increased in UVB-damaged leaves due to the cytoplasmic accumulation of fragmented, depolarized mitochondria. Furthermore, we observed that autophagy was involved in the removal of depolarized mitochondria when mitochondrial function was disrupted by mutation of the FRIENDLY gene, which is required for proper mitochondrial distribution. Therefore, autophagy of mitochondria functions in response to mitochondrion-specific dysfunction as well as UVB damage. Together, these results indicate that autophagy is centrally involved in mitochondrial quality control in Arabidopsis leaves.
Keywords: Arabidopsis (Arabidopsis thaliana); Autophagy; Mitochondria; Mitophagy; Organelle quality control; Ultraviolet B.
� The Author(s) 2020. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists.
Figures





Comment in
-
Mitophagy-A New Player in UV-B Damage Recovery in Plants.Plant Cell Physiol. 2021 May 11;62(2):226-228. doi: 10.1093/pcp/pcab012. Plant Cell Physiol. 2021. PMID: 33515260 No abstract available.
References
-
- Boesch P., Weber-Lotfi F., Ibrahim N., Tarasenko V., Cosset A., Paulus F., et al. (2011) DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim. Biophys. Acta 1813: 186–200. - PubMed
-
- Chung T., Phillips A.R., Vierstra R.D. (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62: 483–493. - PubMed
-
- Clausen C., Ilkavets I., Thomson R., Philippar K., Vojta A., M�Hlmann T., et al. (2004) Intracellular localization of VDAC proteins in plants. Planta 220: 30–37. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

