High-Temporal-Resolution smFISH Method for Gene Expression Studies in Caenorhabditis elegans Embryos
- PMID: 33355449
- PMCID: PMC10619480
- DOI: 10.1021/acs.analchem.0c02966
High-Temporal-Resolution smFISH Method for Gene Expression Studies in Caenorhabditis elegans Embryos
Abstract
Recent development in fluorescence-based molecular tools has contributed significantly to developmental studies, including embryogenesis. Many of these tools rely on multiple steps of sample manipulation, so obtaining large sample sizes presents a major challenge as it can be labor-intensive and time-consuming. However, large sample sizes are required to uncover critical aspects of embryogenesis, for example, subtle phenotypic differences or gene expression dynamics. This problem is particularly relevant for single-molecule fluorescence in situ hybridization (smFISH) studies in Caenorhabditis elegans embryogenesis. Microfluidics can help address this issue by allowing a large number of samples and parallelization of experiments. However, performing efficient reagent exchange on chip for large numbers of embryos remains a bottleneck. Here, we present a microfluidic pipeline for large-scale smFISH imaging of C. elegans embryos with minimized labor. We designed embryo traps and engineered a protocol allowing for efficient chemical exchange for hundreds of C. elegans embryos simultaneously. Furthermore, the device design and small footprint optimize imaging throughput by facilitating spatial registration and enabling minimal user input. We conducted the smFISH protocol on chip and demonstrated that image quality is preserved. With one device replacing the equivalent of 10 glass slides of embryos mounted manually, our microfluidic approach greatly increases throughput. Finally, to highlight the capability of our platform to perform longitudinal studies with high temporal resolution, we conducted a temporal analysis of par-1 gene expression in early C. elegans embryos. The method demonstrated here paves the way for systematic high-temporal-resolution studies that will benefit large-scale RNAi and drug screens and in systems beyond C. elegans embryos.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


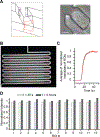
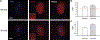
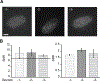

References
-
- Wood WB The Nematode Caenorhabditis Elegans; Cold Spring Harbor Laboratory: New York, 1988.
-
- Bryson-Richardson R; Berger S; Currie P Atlas of Zebrafish Development; Academic Press: San Diego, 2012.
-
- Bate M; Martinez Arias A The Development of Drosophila melanogaster; Cold Spring Harbor Laboratory: New York, 1993.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

