At the Root of T2R Gene Evolution: Recognition Profiles of Coelacanth and Zebrafish Bitter Receptors
- PMID: 33355666
- PMCID: PMC7851594
- DOI: 10.1093/gbe/evaa264
At the Root of T2R Gene Evolution: Recognition Profiles of Coelacanth and Zebrafish Bitter Receptors
Abstract
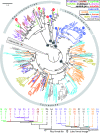
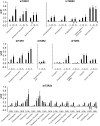
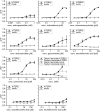
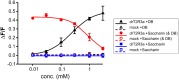
The careful evaluation of food is important for survival throughout the animal kingdom, and specialized chemoreceptors have evolved to recognize nutrients, minerals, acids, and many toxins. Vertebrate bitter taste, mediated by the taste receptor type 2 (T2R) family, warns against potentially toxic compounds. During evolution T2R receptors appear first in bony fish, but the functional properties of bony fish T2R receptors are mostly unknown. We performed a phylogenetic analysis showing the "living fossil" coelacanth (Latimeria chalumnae) and zebrafish (Danio rerio) to possess T2R repertoires typical for early-diverged species in the lobe-finned and the ray-finned clade, respectively. Receptors from these two species were selected for heterologous expression assays using a diverse panel of bitter substances. Remarkably, the ligand profile of the most basal coelacanth receptor, T2R01, is identical to that of its ortholog in zebrafish, consistent with functional conservation across >400 Myr of separate evolution. The second coelacanth receptor deorphaned, T2R02, is activated by steroid hormones and bile acids, evolutionary old molecules that are potentially endogenously synthesized agonists for extraoral T2Rs. For zebrafish, we report the presence of both specialized and promiscuous T2R receptors. Moreover, we identified an antagonist for one of the zebrafish receptors suggesting that bitter antagonism contributed to shape this receptor family throughout evolution.
Keywords: bitter taste receptor; bony fish; calcium mobilization assay.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
References
-
- Adler E, et al. 2000. A novel family of mammalian taste receptors. Cell 100(6):693–702. - PubMed
-
- Ammon C, Schafer J, Kreuzer OJ, Meyerhof W.. 2002. Presence of a plasma membrane targeting sequence in the amino-terminal region of the rat somatostatin receptor 3. Arch Physiol Biochem. 110(1–2):137–145. - PubMed
-
- Ballesteros J, Weinstein H.. 1995. Integrated methods for the construction of three-dimensional models of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25:366–428.
-
- Behrens M, Blank K, Meyerhof W.. 2017. Blends of non-caloric sweeteners saccharin and cyclamate show reduced off-taste due to TAS2R bitter receptor inhibition. Cell Chem Biol. 24(10):1199–1204 e1192. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

