Massively parallel functional testing of MSH2 missense variants conferring Lynch syndrome risk
- PMID: 33357406
- PMCID: PMC7820803
- DOI: 10.1016/j.ajhg.2020.12.003
Massively parallel functional testing of MSH2 missense variants conferring Lynch syndrome risk
Abstract
The lack of functional evidence for the majority of missense variants limits their clinical interpretability and poses a key barrier to the broad utility of carrier screening. In Lynch syndrome (LS), one of the most highly prevalent cancer syndromes, nearly 90% of clinically observed missense variants are deemed "variants of uncertain significance" (VUS). To systematically resolve their functional status, we performed a massively parallel screen in human cells to identify loss-of-function missense variants in the key DNA mismatch repair factor MSH2. The resulting functional effect map is substantially complete, covering 94% of the 17,746 possible variants, and is highly concordant (96%) with existing functional data and expert clinicians' interpretations. The large majority (89%) of missense variants were functionally neutral, perhaps unexpectedly in light of its evolutionary conservation. These data provide ready-to-use functional evidence to resolve the ∼1,300 extant missense VUSs in MSH2 and may facilitate the prospective classification of newly discovered variants in the clinic.
Keywords: DNA mismatch repair; Lynch syndrome; MSH2; cancer; deep mutational scanning; genotype-phenotype; variants of uncertain significance.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

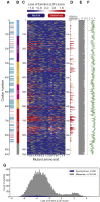

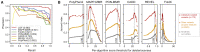

References
-
- Lynch H.T., Krush A.J. Cancer family “G” revisited: 1895-1970. Cancer. 1971;27:1505–1511. - PubMed
-
- Leach F.S., Nicolaides N.C., Papadopoulos N., Liu B., Jen J., Parsons R., Peltomäki P., Sistonen P., Aaltonen L.A., Nyström-Lahti M. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. - PubMed
-
- Fishel R., Lescoe M.K., Rao M.R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1994;77 1, 166. - PubMed
-
- Bronner C.E., Baker S.M., Morrison P.T., Warren G., Smith L.G., Lescoe M.K., Kane M., Earabino C., Lipford J., Lindblom A. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. - PubMed
-
- Papadopoulos N., Nicolaides N.C., Wei Y.F., Ruben S.M., Carter K.C., Rosen C.A., Haseltine W.A., Fleischmann R.D., Fraser C.M., Adams M.D. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

