RTEL1 Regulates G4/R-Loops to Avert Replication-Transcription Collisions
- PMID: 33357438
- PMCID: PMC7773548
- DOI: 10.1016/j.celrep.2020.108546
RTEL1 Regulates G4/R-Loops to Avert Replication-Transcription Collisions
Abstract
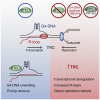
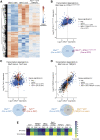
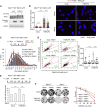
Regulator of telomere length 1 (RTEL1) is an essential helicase that maintains telomere integrity and facilitates DNA replication. The source of replication stress in Rtel1-deficient cells remains unclear. Here, we report that loss of RTEL1 confers extensive transcriptional changes independent of its roles at telomeres. The majority of affected genes in Rtel1-/- cells possess G-quadruplex (G4)-DNA-forming sequences in their promoters and are similarly altered at a transcriptional level in wild-type cells treated with the G4-DNA stabilizer TMPyP4 (5,10,15,20-Tetrakis-(N-methyl-4-pyridyl)porphine). Failure to resolve G4-DNAs formed in the displaced strand of RNA-DNA hybrids in Rtel1-/- cells is suggested by increased R-loops and elevated transcription-replication collisions (TRCs). Moreover, removal of R-loops by RNaseH1 overexpression suppresses TRCs and alleviates the global replication defects observed in Rtel1-/- and Rtel1PIP_box knockin cells and in wild-type cells treated with TMPyP4. We propose that RTEL1 unwinds G4-DNA/R-loops to avert TRCs, which is important to prevent global deregulation in both transcription and DNA replication.
Keywords: G-quadruplexes; G4-DNA structures; R-loops; RTEL1; genome instability; replication stress; transcription.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests S.J.B. is also scientific co-founder and VP Science Strategy at Artios Pharma, Babraham Research Campus, Cambridge, UK.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

