A Primer on Dose-Response Data Modeling in Radiation Therapy
- PMID: 33358230
- PMCID: PMC9339232
- DOI: 10.1016/j.ijrobp.2020.11.020
A Primer on Dose-Response Data Modeling in Radiation Therapy
Abstract
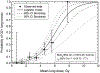
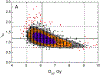
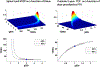
An overview of common approaches used to assess a dose response for radiation therapy-associated endpoints is presented, using lung toxicity data sets analyzed as a part of the High Dose per Fraction, Hypofractionated Treatment Effects in the Clinic effort as an example. Each component presented (eg, data-driven analysis, dose-response analysis, and calculating uncertainties on model prediction) is addressed using established approaches. Specifically, the maximum likelihood method was used to calculate best parameter values of the commonly used logistic model, the profile-likelihood to calculate confidence intervals on model parameters, and the likelihood ratio to determine whether the observed data fit is statistically significant. The bootstrap method was used to calculate confidence intervals for model predictions. Correlated behavior of model parameters and implication for interpreting dose response are discussed.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest:
Figures





Comment in
-
In Regard to Moiseenko et al.Int J Radiat Oncol Biol Phys. 2021 Jun 1;110(2):609. doi: 10.1016/j.ijrobp.2021.03.018. Int J Radiat Oncol Biol Phys. 2021. PMID: 33989579 No abstract available.
-
In Reply to Schultheiss.Int J Radiat Oncol Biol Phys. 2021 Aug 1;110(5):1541-1543. doi: 10.1016/j.ijrobp.2021.03.019. Epub 2021 May 18. Int J Radiat Oncol Biol Phys. 2021. PMID: 34024669 No abstract available.
References
-
- Bentzen SM, Tucker SL. Quantifying the position and steepness of radiation dose-response curves. Int J Radiat Biol 1997; 71:531–42. doi. - PubMed
-
- Roberts SA, Hendry JH. The delay before onset of accelerated tumour cell repopulation during radiotherapy: A direct maximum-likelihood analysis of a collection of worldwide tumour-control data. Radiother Oncol 1993; 29:69–74. doi. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

