SARS-Cov-2 ORF3a: Mutability and function
- PMID: 33359807
- PMCID: PMC7836370
- DOI: 10.1016/j.ijbiomac.2020.12.142
SARS-Cov-2 ORF3a: Mutability and function
Abstract
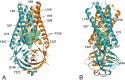
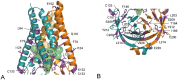
In this study, analysis of changes of SARS-CoV-2 ORF3a protein during pandemic is reported. ORF3a, a conserved coronavirus protein, is involved in virus replication and release. A set of 70,752 high-quality SARS-CoV-2 genomes available in GISAID databank at the end of August 2020 have been scanned. All ORF3a mutations in the virus genomes were grouped according to the collection date interval and over the entire data set. The considered intervals were: start of collection-February, March, April, May, June, July and August 2020. The top five most frequent variants were examined within each collection interval. Overall, seventeen variants have been isolated. Ten of the seventeen mutant sites occur within the transmembrane (TM) domain of ORF3a and are in contact with the central pore or side tunnels. The other variant sites are in different places of the ORF3a structure. Within the entire sample, the five most frequent mutations are V13L, Q57H, Q57H + A99V, G196V and G252V. The same analysis identified 28 sites identically conserved in all the genome isolates. These sites are possibly involved in stabilization of monomer, dimer, tetramerization and interaction with other cellular components. The results here reported can be helpful to understand virus biology and to design new therapeutic strategies.
Keywords: Conserved sites; Mutated sites; ORF3a; Pore; Q57H; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest None.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

