Profiling of immune dysfunction in COVID-19 patients allows early prediction of disease progression
- PMID: 33361110
- PMCID: PMC7768198
- DOI: 10.26508/lsa.202000955
Profiling of immune dysfunction in COVID-19 patients allows early prediction of disease progression
Abstract
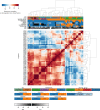
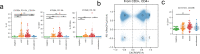
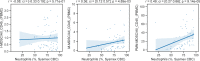
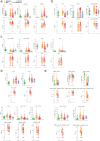
With a rising incidence of COVID-19-associated morbidity and mortality worldwide, it is critical to elucidate the innate and adaptive immune responses that drive disease severity. We performed longitudinal immune profiling of peripheral blood mononuclear cells from 45 patients and healthy donors. We observed a dynamic immune landscape of innate and adaptive immune cells in disease progression and absolute changes of lymphocyte and myeloid cells in severe versus mild cases or healthy controls. Intubation and death were coupled with selected natural killer cell KIR receptor usage and IgM+ B cells and associated with profound CD4 and CD8 T-cell exhaustion. Pseudo-temporal reconstruction of the hierarchy of disease progression revealed dynamic time changes in the global population recapitulating individual patients and the development of an eight-marker classifier of disease severity. Estimating the effect of clinical progression on the immune response and early assessment of disease progression risks may allow implementation of tailored therapies.
© 2020 Rendeiro et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures









Update of
-
Longitudinal immune profiling of mild and severe COVID-19 reveals innate and adaptive immune dysfunction and provides an early prediction tool for clinical progression.medRxiv [Preprint]. 2020 Sep 9:2020.09.08.20189092. doi: 10.1101/2020.09.08.20189092. medRxiv. 2020. Update in: Life Sci Alliance. 2020 Dec 24;4(2):e202000955. doi: 10.26508/lsa.202000955. PMID: 32935114 Free PMC article. Updated. Preprint.
References
-
- Agrati C, Sacchi A, Bordoni V, Cimini E, Notari S, Grassi G, Casetti R, Tartaglia E, Lalle E, D’Abramo A, et al. (2020) Expansion of myeloid-derived suppressor cells in patients with severe coronavirus disease (COVID-19). Cell Death Differ 27: 3196–3207. 10.1038/s41418-020-0572-6 - DOI - PMC - PubMed
-
- Aschenbrenner AC, Mouktaroudi M, Kraemer B, Antonakos N, Oestreich M, Gkizeli K, Nuesch-Germano M, Saridaki M, Bonaguro L, Reusch N, et al. (2020) Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. BioRxiv–Infectious Diseases (except HIV/AIDS) 10.1101/2020.07.07.20148395 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
