Protein dosage of the lldPRD operon is correlated with RNase E-dependent mRNA processing
- PMID: 33361194
- PMCID: PMC8095457
- DOI: 10.1128/JB.00555-20
Protein dosage of the lldPRD operon is correlated with RNase E-dependent mRNA processing
Abstract
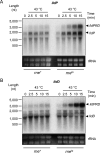
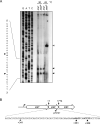
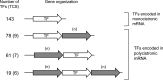
The ability of Escherichia coli to grow on L-lactate as a sole carbon source depends on the expression of the lldPRD operon. A striking feature of this operon is that the transcriptional regulator (LldR) encoding gene is located between the permease (LldP) and the dehydrogenase (LldD) encoding genes. In this study we report that dosage of the LldP, LldR, and LldD proteins is not modulated on the transcriptional level. Instead, modulation of protein dosage is primarily correlated with RNase E-dependent mRNA processing events that take place within the lldR mRNA, leading to the immediate inactivation of lldR, to differential segmental stabilities of the resulting cleavage products, and to differences in the translation efficiencies of the three cistrons. A model for the processing events controlling the molar quantities of the proteins in the lldPRD operon is presented and discussed.ImportanceAdjustment of gene expression is critical for proper cell function. For the case of polycistronic transcripts, posttranscriptional regulatory mechanisms can be used to fine-tune the expression of individual cistrons. Here, we elucidate how protein dosage of the Escherichia coli lldPRD operon, which presents the paradox of having the gene encoding a regulator protein located between genes that code for a permease and an enzyme, is regulated. Our results demonstrate that the key event in this regulatory mechanism involves the RNase E-dependent cleavage of the primary lldPRD transcript at internal site(s) located within the lldR cistron, resulting in a drastic decrease of intact lldR mRNA, to differential segmental stabilities of the resulting cleavage products, and to differences in the translation efficiencies of the three cistrons.
Copyright © 2020 American Society for Microbiology.
Figures



Similar articles
-
Dual role of LldR in regulation of the lldPRD operon, involved in L-lactate metabolism in Escherichia coli.J Bacteriol. 2008 Apr;190(8):2997-3005. doi: 10.1128/JB.02013-07. Epub 2008 Feb 8. J Bacteriol. 2008. PMID: 18263722 Free PMC article.
-
Expanded roles of lactate-sensing LldR in transcription regulation of the Escherichia coli K-12 genome: lactate utilisation and acid resistance.Microb Genom. 2023 May;9(5):mgen001015. doi: 10.1099/mgen.0.001015. Microb Genom. 2023. PMID: 37219924 Free PMC article.
-
Lactate utilization is regulated by the FadR-type regulator LldR in Pseudomonas aeruginosa.J Bacteriol. 2012 May;194(10):2687-92. doi: 10.1128/JB.06579-11. Epub 2012 Mar 9. J Bacteriol. 2012. PMID: 22408166 Free PMC article.
-
The cyanase operon and cyanate metabolism.FEMS Microbiol Rev. 1990 Dec;7(3-4):247-52. doi: 10.1111/j.1574-6968.1990.tb04920.x. FEMS Microbiol Rev. 1990. PMID: 2094285 Review.
-
Post-transcriptional control in the polycistronic operon environment: studies of the atp operon of Escherichia coli.Mol Microbiol. 1990 Aug;4(8):1233-40. doi: 10.1111/j.1365-2958.1990.tb00702.x. Mol Microbiol. 1990. PMID: 2149159 Review.
References
-
- Futai M, Kimura H. 1977. Inducible membrane-bound l-lactate dehydrogenase from Escherichia coli. J Biol Chem 252:5820–5827. - PubMed
-
- Nishimura Y, Tan IKP, Ohgami Y, Kohgami K, Kamihara T. 1983. Induction of membrane-bound l-lactate dehydrogenase in Escherichia coli under conditions of nitrate respiration, fumarate reduction and trimethylamine-N-oxide reduction. FEMS Microbiol Lett 17:283–286.
LinkOut - more resources
Full Text Sources
Other Literature Sources

