A High-Throughput Method for Screening for Genes Controlling Bacterial Conjugation of Antibiotic Resistance
- PMID: 33361328
- PMCID: PMC7762799
- DOI: 10.1128/mSystems.01226-20
A High-Throughput Method for Screening for Genes Controlling Bacterial Conjugation of Antibiotic Resistance
Abstract
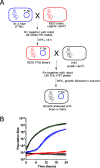
The rapid horizontal transmission of antibiotic resistance genes on conjugative plasmids between bacterial host cells is a major cause of the accelerating antibiotic resistance crisis. There are currently no experimental platforms for fast and cost-efficient screening of genetic effects on antibiotic resistance transmission by conjugation, which prevents understanding and targeting conjugation. We introduce a novel experimental framework to screen for conjugation-based horizontal transmission of antibiotic resistance between >60,000 pairs of cell populations in parallel. Plasmid-carrying donor strains are constructed in high-throughput. We then mix the resistance plasmid-carrying donors with recipients in a design where only transconjugants can reproduce, measure growth in dense intervals, and extract transmission times as the growth lag. As proof-of-principle, we exhaustively explore chromosomal genes controlling F-plasmid donation within Escherichia coli populations, by screening the Keio deletion collection in high replication. We recover all seven known chromosomal gene mutants affecting conjugation as donors and identify many novel mutants, all of which diminish antibiotic resistance transmission. We validate nine of the novel genes' effects in liquid mating assays and complement one of the novel genes' effect on conjugation (rseA). The new framework holds great potential for exhaustive disclosing of candidate targets for helper drugs that delay resistance development in patients and societies and improve the longevity of current and future antibiotics. Further, the platform can easily be adapted to explore interspecies conjugation, plasmid-borne factors, and experimental evolution and be used for rapid construction of strains.IMPORTANCE The rapid transmission of antibiotic resistance genes on conjugative plasmids between bacterial host cells is a major cause of the accelerating antibiotic resistance crisis. There are currently no experimental platforms for fast and cost-efficient screening of genetic effects on antibiotic resistance transmission by conjugation, which prevents understanding and targeting conjugation. We introduce a novel experimental framework to screen for conjugation-based horizontal transmission of antibiotic resistance between >60,000 pairs of cell populations in parallel. As proof-of-principle, we exhaustively explore chromosomal genes controlling F-plasmid donation within E. coli populations. We recover all previously known and many novel chromosomal gene mutants that affect conjugation efficiency. The new framework holds great potential for rapid screening of compounds that decrease transmission. Further, the platform can easily be adapted to explore interspecies conjugation, plasmid-borne factors, and experimental evolution and be used for rapid construction of strains.
Keywords: Escherichia coli; antibiotic resistance; conjugation; high-throughput screening; horizontal gene transfer; horizontal transmission; plasmid.
Copyright © 2020 Alalam et al.
Figures


References
-
- WHO. 2014. Antimicrobial resistance: global report on surveillance 2014. WHO, Geneva, Switzerland.
-
- Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. - DOI - PubMed
-
- WHO. 2017. Report on antibacterial agents in clinical development. WHO, Geneva, Switzerland.
-
- The Pew Charitable Trusts. 2020. Tracking the global pipeline of antibiotics in development, April 2020. The Pew Charitable Trusts, Washington, DC.
LinkOut - more resources
Full Text Sources
Research Materials

