Phosphatidylinositol 3-Phosphate Mediates the Establishment of Infectious Bursal Disease Virus Replication Complexes in Association with Early Endosomes
- PMID: 33361427
- PMCID: PMC8094959
- DOI: 10.1128/JVI.02313-20
Phosphatidylinositol 3-Phosphate Mediates the Establishment of Infectious Bursal Disease Virus Replication Complexes in Association with Early Endosomes
Abstract
Keywords: birnavirus; double-stranded RNA virus; endosomes; phosphoinositides; viral replication.
Copyright © 2021 American Society for Microbiology.
Figures




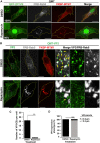



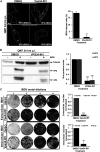

Similar articles
-
On the role of VP3-PI3P interaction in birnavirus endosomal membrane targeting.Elife. 2025 Mar 6;13:RP97261. doi: 10.7554/eLife.97261. Elife. 2025. PMID: 40047543 Free PMC article.
-
The Formation and Function of Birnaviridae Virus Factories.Int J Mol Sci. 2023 May 9;24(10):8471. doi: 10.3390/ijms24108471. Int J Mol Sci. 2023. PMID: 37239817 Free PMC article. Review.
-
Infectious Bursal Disease Virus Hijacks Endosomal Membranes as the Scaffolding Structure for Viral Replication.J Virol. 2018 May 14;92(11):e01964-17. doi: 10.1128/JVI.01964-17. Print 2018 Jun 1. J Virol. 2018. PMID: 29540593 Free PMC article.
-
Rab1b-GBF1-ARF1 Secretory Pathway Axis Is Required for Birnavirus Replication.J Virol. 2022 Feb 23;96(4):e0200521. doi: 10.1128/JVI.02005-21. Epub 2021 Dec 8. J Virol. 2022. PMID: 34878889 Free PMC article.
-
Role of MicroRNAs in Host Defense against Infectious Bursal Disease Virus (IBDV) Infection: A Hidden Front Line.Viruses. 2020 May 14;12(5):543. doi: 10.3390/v12050543. Viruses. 2020. PMID: 32423052 Free PMC article. Review.
Cited by
-
CAPRIN1 Is Required for Control of Viral Replication Complexes by Interferon Gamma.mBio. 2023 Jun 27;14(3):e0017223. doi: 10.1128/mbio.00172-23. Epub 2023 Apr 13. mBio. 2023. PMID: 37052473 Free PMC article.
-
Infectious Bursal Disease Virus Assembly Causes Endoplasmic Reticulum Stress and Lipid Droplet Accumulation.Viruses. 2023 May 31;15(6):1295. doi: 10.3390/v15061295. Viruses. 2023. PMID: 37376595 Free PMC article.
-
Chicken Heat Shock Protein 70 Is an Essential Host Protein for Infectious Bursal Disease Virus Infection In Vitro.Pathogens. 2021 May 28;10(6):664. doi: 10.3390/pathogens10060664. Pathogens. 2021. PMID: 34071696 Free PMC article.
-
On the role of VP3-PI3P interaction in birnavirus endosomal membrane targeting.Elife. 2025 Mar 6;13:RP97261. doi: 10.7554/eLife.97261. Elife. 2025. PMID: 40047543 Free PMC article.
-
The Formation and Function of Birnaviridae Virus Factories.Int J Mol Sci. 2023 May 9;24(10):8471. doi: 10.3390/ijms24108471. Int J Mol Sci. 2023. PMID: 37239817 Free PMC article. Review.
References
-
- Cosgrove AS. 1962. An apparently new disease of chickens: avian nephrosis. Avian Dis 6:385–389. doi:10.2307/1587909. - DOI
-
- Luque D, Mata CP, González-Camacho F, González JM, Gómez-Blanco J, Alfonso C, Rivas G, Havens WM, Kanematsu S, Suzuki N, Ghabrial SA, Trus BL, Castón JR. 2016. Heterodimers as the structural unit of the T=1 capsid of the fungal double-stranded RNA Rosellinia necatrix quadrivirus 1. J Virol 90:11220–11230. doi:10.1128/JVI.01013-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

