Chromothripsis drives the evolution of gene amplification in cancer
- PMID: 33361815
- PMCID: PMC7933129
- DOI: 10.1038/s41586-020-03064-z
Chromothripsis drives the evolution of gene amplification in cancer
Erratum in
-
Publisher Correction: Chromothripsis drives the evolution of gene amplification in cancer.Nature. 2021 Mar;591(7850):E19. doi: 10.1038/s41586-021-03379-5. Nature. 2021. PMID: 33649505 No abstract available.
Abstract
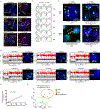
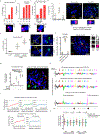
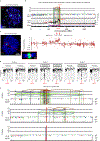
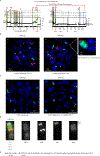
Focal chromosomal amplification contributes to the initiation of cancer by mediating overexpression of oncogenes1-3, and to the development of cancer therapy resistance by increasing the expression of genes whose action diminishes the efficacy of anti-cancer drugs. Here we used whole-genome sequencing of clonal cell isolates that developed chemotherapeutic resistance to show that chromothripsis is a major driver of circular extrachromosomal DNA (ecDNA) amplification (also known as double minutes) through mechanisms that depend on poly(ADP-ribose) polymerases (PARP) and the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs). Longitudinal analyses revealed that a further increase in drug tolerance is achieved by structural evolution of ecDNAs through additional rounds of chromothripsis. In situ Hi-C sequencing showed that ecDNAs preferentially tether near chromosome ends, where they re-integrate when DNA damage is present. Intrachromosomal amplifications that formed initially under low-level drug selection underwent continuing breakage-fusion-bridge cycles, generating amplicons more than 100 megabases in length that became trapped within interphase bridges and then shattered, thereby producing micronuclei whose encapsulated ecDNAs are substrates for chromothripsis. We identified similar genome rearrangement profiles linked to localized gene amplification in human cancers with acquired drug resistance or oncogene amplifications. We propose that chromothripsis is a primary mechanism that accelerates genomic DNA rearrangement and amplification into ecDNA and enables rapid acquisition of tolerance to altered growth conditions.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures










Comment in
-
No pains, no gains: how chromosome fragmentation promotes gene amplification.Mol Cell. 2021 Mar 4;81(5):901-904. doi: 10.1016/j.molcel.2021.02.018. Mol Cell. 2021. PMID: 33667381
References
-
- Benner SE, Wahl GM & Von Hoff DD Double minute chromosomes and homogeneously staining regions in tumors taken directly from patients versus in human tumor cell lines. Anticancer Drugs 2, 11–25 (1991). - PubMed
-
- Alt FW, Kellems RE, Bertino JR & Schimke RT Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem 253, 1357–1370 (1978). - PubMed
-
- Kaufman RJ, Brown PC & Schimke RT Amplified dihydrofolate reductase genes in unstably methotrexate-resistant cells are associated with double minute chromosomes. Proc Natl Acad Sci U S A 76, 5669–5673 (1979).
References for Methods section
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

