A Mass Spectrometry Based Metabolite Profiling Workflow for Selecting Abundant Specific Markers and Their Structurally Related Multi-Component Signatures in Traditional Chinese Medicine Multi-Herb Formulae
- PMID: 33362543
- PMCID: PMC7756971
- DOI: 10.3389/fphar.2020.578346
A Mass Spectrometry Based Metabolite Profiling Workflow for Selecting Abundant Specific Markers and Their Structurally Related Multi-Component Signatures in Traditional Chinese Medicine Multi-Herb Formulae
Abstract
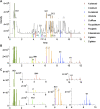
In Traditional Chinese Medicine (TCM), herbal preparations often consist of a mixture of herbs. Their quality control is challenging because every single herb contains hundreds of components (secondary metabolites). A typical 10 herb TCM formula was selected to develop an innovative strategy for its comprehensive chemical characterization and to study the specific contribution of each herb to the formula in an exploratory manner. Metabolite profiling of the TCM formula and the extract of each single herb were acquired with liquid chromatography coupled to high-resolution mass spectrometry for qualitative analyses, and to evaporative light scattering detection (ELSD) for semi-quantitative evaluation. The acquired data were organized as a feature-based molecular network (FBMN) which provided a comprehensive view of all types of secondary metabolites and their occurrence in the formula and all single herbs. These features were annotated by combining MS/MS-based in silico spectral match, manual evaluation of the structural consistency in the FBMN clusters, and taxonomy information. ELSD detection was used as a filter to select the most abundant features. At least one marker per herb was highlighted based on its specificity and abundance. A single large-scale fractionation from the enriched formula enabled the isolation and formal identification of most of them. The obtained markers allowed an improved annotation of associated features by manually propagating this information through the FBMN. These data were incorporated in the high-resolution metabolite profiling of the formula, which highlighted specific series of related components to each individual herb markers. These series of components, named multi-component signatures, may serve to improve the traceability of each herb in the formula. Altogether, the strategy provided highly informative compositional data of the TCM formula and detailed visualizations of the contribution of each herb by FBMN, filtered feature maps, and reconstituted chromatogram traces of all components linked to each specific marker. This comprehensive MS-based analytical workflow allowed a generic and unbiased selection of specific and abundant markers and the identification of multiple related sub-markers. This exploratory approach could serve as a starting point to develop more simple and targeted quality control methods with adapted marker specificity selection criteria to given TCM formula.
Keywords: Traditional Chinese Medicine; feature-based molecular network; multi-component signature, TCM, Mass spectrometry, Chemical markers; multi-herb formulae; quality control.
Copyright © 2020 Houriet, Allard, Queiroz, Marcourt, Gaudry, Vallin, Li, Lin, Wang, Kuchta and Wolfender.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Afzan A., Wolfender J.-L., Afzan A., Kasim N., Ismail Nor H., Azmi N., et al. (2019). Differentiation of Ficus deltoidea varieties and chemical marker determination by UHPLC-TOFMS metabolomics for establishing quality control criteria of this popular Malaysian medicinal herb. Metabolomics 15 (3), 35 10.1007/s11306-019-1489-2 - DOI - PubMed
LinkOut - more resources
Full Text Sources

