Disentangling Biomolecular Corona Interactions With Cell Receptors and Implications for Targeting of Nanomedicines
- PMID: 33363128
- PMCID: PMC7758247
- DOI: 10.3389/fbioe.2020.599454
Disentangling Biomolecular Corona Interactions With Cell Receptors and Implications for Targeting of Nanomedicines
Abstract
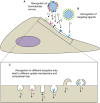
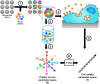
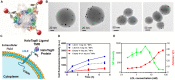
Nanoparticles are promising tools for nanomedicine in a wide array of therapeutic and diagnostic applications. Yet, despite the advances in the biomedical applications of nanomaterials, relatively few nanomedicines made it to the clinics. The formation of the biomolecular corona on the surface of nanoparticles has been known as one of the challenges toward successful targeting of nanomedicines. This adsorbed protein layer can mask targeting moieties and creates a new biological identity that critically affects the subsequent biological interactions of nanomedicines with cells. Extensive studies have been directed toward understanding the characteristics of this layer of biomolecules and its implications for nanomedicine outcomes at cell and organism levels, yet several aspects are still poorly understood. One aspect that still requires further insights is how the biomolecular corona interacts with and is "read" by the cellular machinery. Within this context, this review is focused on the current understanding of the interactions of the biomolecular corona with cell receptors. First, we address the importance and the role of receptors in the uptake of nanoparticles. Second, we discuss the recent advances and techniques in characterizing and identifying biomolecular corona-receptor interactions. Additionally, we present how we can exploit the knowledge of corona-cell receptor interactions to discover novel receptors for targeting of nanocarriers. Finally, we conclude this review with an outlook on possible future perspectives in the field. A better understanding of the first interactions of nanomaterials with cells, and -in particular -the receptors interacting with the biomolecular corona and involved in nanoparticle uptake, will help for the successful design of nanomedicines for targeted delivery.
Keywords: biomolecular corona; cell receptors; corona-receptor interactions; nanoparticles; targeting; uptake.
Copyright © 2020 Aliyandi, Zuhorn and Salvati.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources

