Proteome of the Triatomine Digestive Tract: From Catalytic to Immune Pathways; Focusing on Annexin Expression
- PMID: 33363206
- PMCID: PMC7755933
- DOI: 10.3389/fmolb.2020.589435
Proteome of the Triatomine Digestive Tract: From Catalytic to Immune Pathways; Focusing on Annexin Expression
Abstract
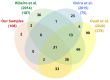
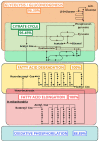
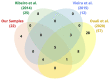
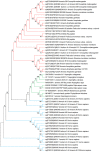
Rhodnius prolixus, Panstrongylus megistus, Triatoma infestans, and Dipetalogaster maxima are all triatomines and potential vectors of the protozoan Trypanosoma cruzi responsible for human Chagas' disease. Considering that the T. cruzi's cycle occurs inside the triatomine digestive tract (TDT), the analysis of the TDT protein profile is an essential step to understand TDT physiology during T. cruzi infection. To characterize the protein profile of TDT of D. maxima, P. megistus, R. prolixus, and T. infestans, a shotgun liquid chromatography-tandem mass spectrometry (LC-MS/MS) approach was applied in this report. Most proteins were found to be closely related to metabolic pathways such as gluconeogenesis/glycolysis, citrate cycle, fatty acid metabolism, oxidative phosphorylation, but also to the immune system. We annotated this new proteome contribution gathering it with those previously published in accordance with Gene Ontology and KEGG. Enzymes were classified in terms of class, acceptor, and function, while the proteins from the immune system were annotated by reference to the pathways of humoral response, cell cycle regulation, Toll, IMD, JNK, Jak-STAT, and MAPK, as available from the Insect Innate Immunity Database (IIID). These pathways were further subclassified in recognition, signaling, response, coagulation, melanization and none. Finally, phylogenetic affinities and gene expression of annexins were investigated for understanding their role in the protection and homeostasis of intestinal epithelial cells against the inflammation.
Keywords: annexin; chagas disease; digestive tract; enzymes; immunity; mass spectrometry; triatomine.
Copyright © 2020 Gumiel, Mattos, Vieira, Moraes, Moreira, Gonzalez, Teixeira-Ferreira, Waghabi, Azambuja and Carels.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Albuquerque-Cunha J. M., Gonzalez M. S., Garcia E. S., Melllo C. B., Azambuja P., Almeida J. C. A., et al. (2009). Cytochemical characterization of microvillar and perimicrovillar membranes in the posterior midgut epithelium of Rhodnius prolixus. Arthropod. Struct. Dev. 38 31–44. 10.1016/j.asd.2008.06.001 - DOI - PubMed
-
- Araújo C. A., Waniek P. J., Stock P., Mayer C., Jansen A. M., Schaub G. A. (2006). Sequence characterization and expression patterns of defensin and lysozyme encoding genes from the gut of the reduviid bug Triatoma brasiliensis. Insect. Biochem. Mol. Biol. 36 547–560. 10.1016/j.ibmb.2006.04.003 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

